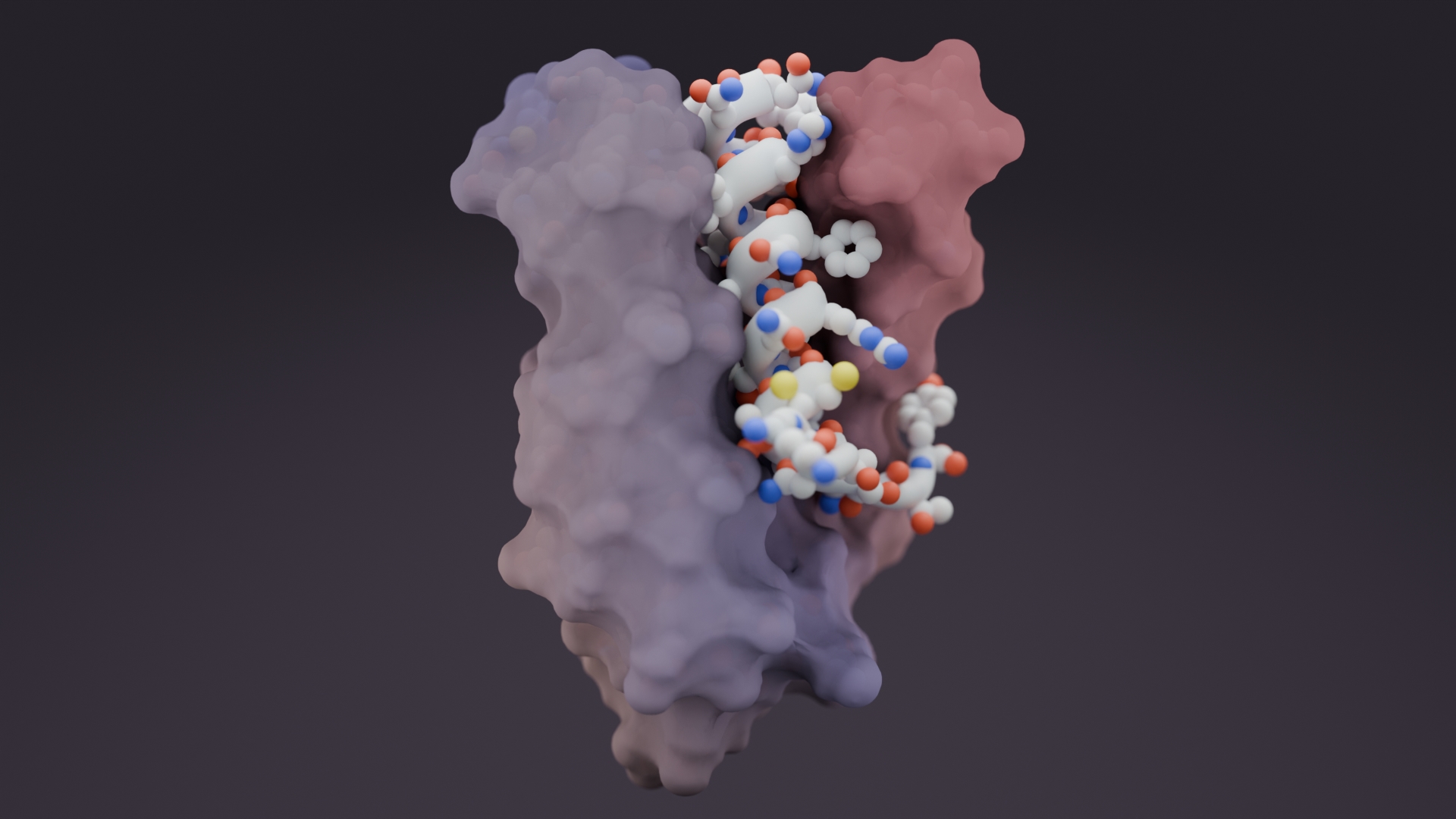
Today, a multidisciplinary team of researchers at the University of Washington, Fred Hutch, and The Scripps Research Institute published in Nature Biotechnology the computational design of a trimeric influenza-neutralizing protein that binds extremely tightly to the H3 hemagglutinin of 1968 Hong Kong pandemic influenza virus (A/Hong Kong/X31/1968). It also cross-reacts with human relevant H1, H2 and H3 influenza strains.
The research has been recognized by opinion leaders and media outlets as a major step in the fight against the flu. See articles in Science , the Conversation, and Scientific American, C&EN News.
Crafted in the Baker lab at the Institute for Protein Design, the protein affectionately known “Flu-Glue” was shown by the Fuller lab at UW to completely protect mice when given as a single intranasal dose 24 h before or after lethal challenge with H3N2 influenza. The Bloom lab at the Fred Hutch has also shown that Flu-Glu has broad specificity to block both H3 and H1 viruses in vitro. Also, Flu-Glue can both capture and detect hemagglutinin in a low cost paper-based diagnostic assay developed in collaboration with the Yager lab at the UW.
How does it work?
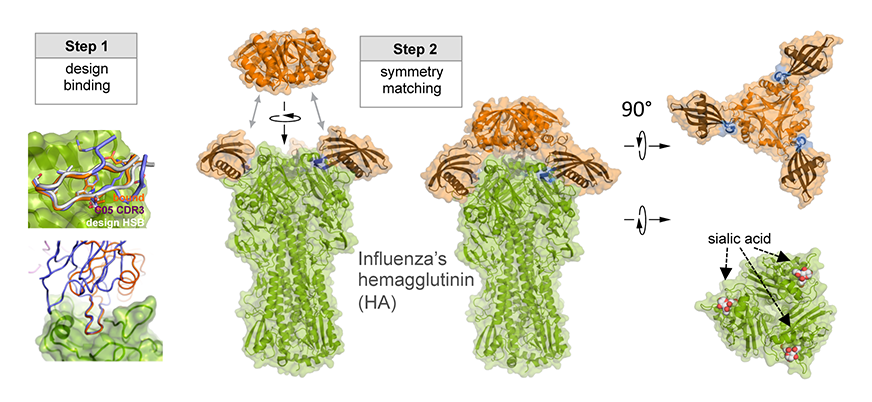
As illustrated in Figure 1, researchers designed this potent protein in a two-stage process. They first used Rosetta computational design algorithms to generate a soluble protein that binds with reasonable affinity to the sialic acid binding pocket of the virus’ hemagglutinin protein (HA). This is the site of receptor binding for virus which enables it to grab onto the surface cells and infect them. In a second step, researchers then went on to design a homo-oligomeric trimeric version of the protein that self-assembles to optimally position three the binding proteins to match with near atomic level accuracy the three sialic acid binding pockets in of the self-assembled HA trimer—this is the natural form of HA on the surface of the virus. By perfectly pre-arranging three low affinity HA binders to match three identical pockets on the surface of HA, the team achieved very tight binding to flu HA. The Wilson lab and Ward lab at TSRI confirmed these structures by X-ray crystallography and cryo electron microscopy.
Why is it important?
Many viruses such as Ebola, influenza, respiratory syncytial virus, and others use a trimeric architecture for their cell surface receptor binding proteins. This work proves that protein design can achieve very tight binding to such viral proteins with prophylactic, therapeutic, and diagnostic application. While it is known that antibodies can bind and neutralize viral receptor proteins, their dimeric architectures are not suited to achieve the exquisite affinity and virus blocking ability of of the computationally designed trimeric Flu-Glue.