Latest posts
-
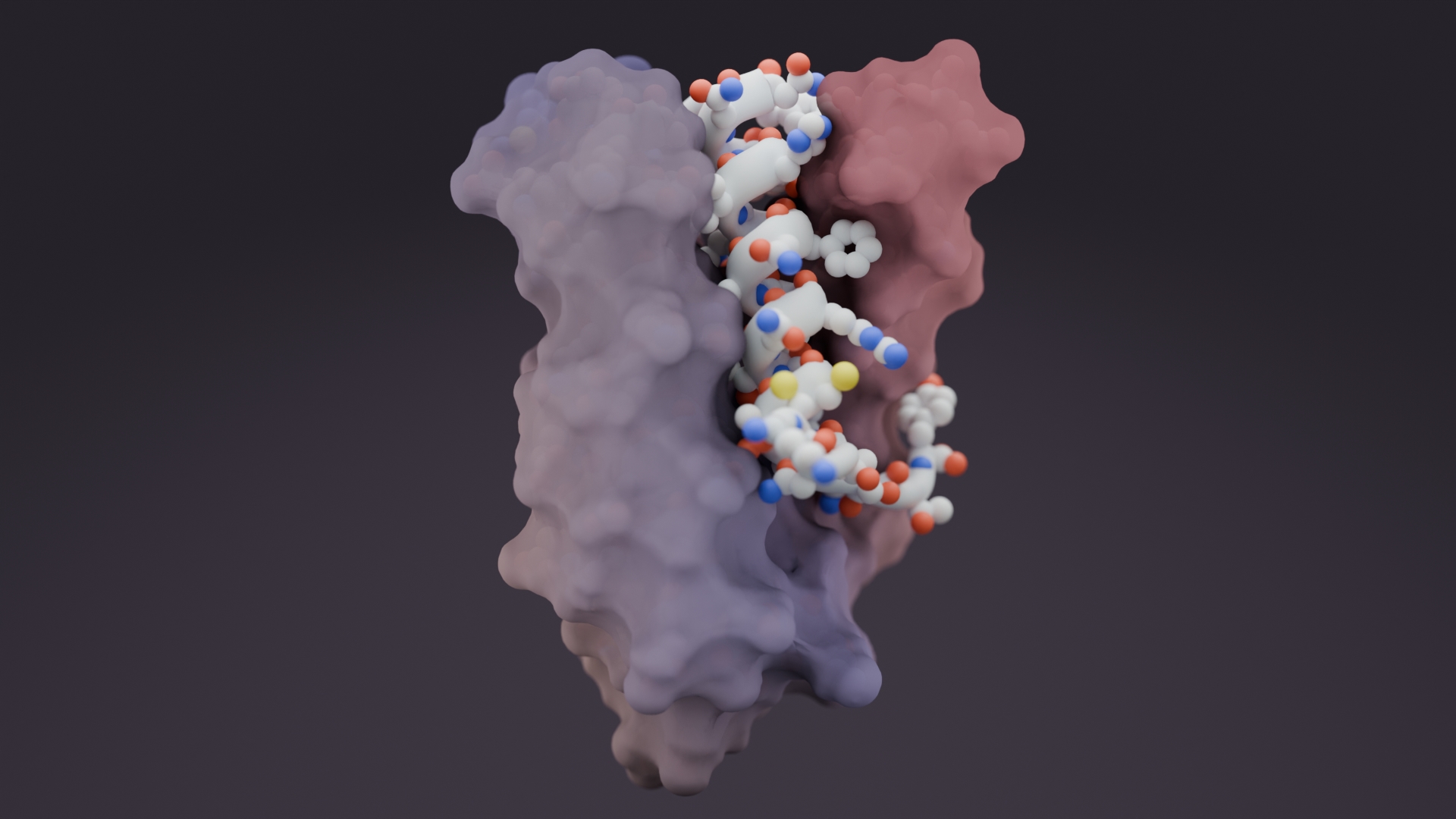
Hitting ‘undruggable’ disease targets
Custom proteins made with AI bind disordered proteins and peptides with atomic precision.
-
Introducing RFdiffusion2
A new deep-learning model developed by scientists at the IPD and MIT is redefining what’s possible in enzyme design.
-
NASEM Releases Consensus Report on Biosecurity and AI
By understanding the opportunities and risks of AI advances, we can collectively advance science in ways that improve the health of all people.
-
Introducing LigandMPNN
ProteinMPNN revolutionized protein sequence design. LigandMPNN opens the door to programming protein–ligand interfaces of all kinds.
-
Neutralizing deadly snake toxins
New AI-generated proteins that protect animals in the lab promise safer, more accessible antivenoms.
-
Derrick Hicks wins additional WRF translational funding
Funding will enable further development of Hicks’ minibinder conjugates as an improved option for radiotherapy cancer treatments.
-
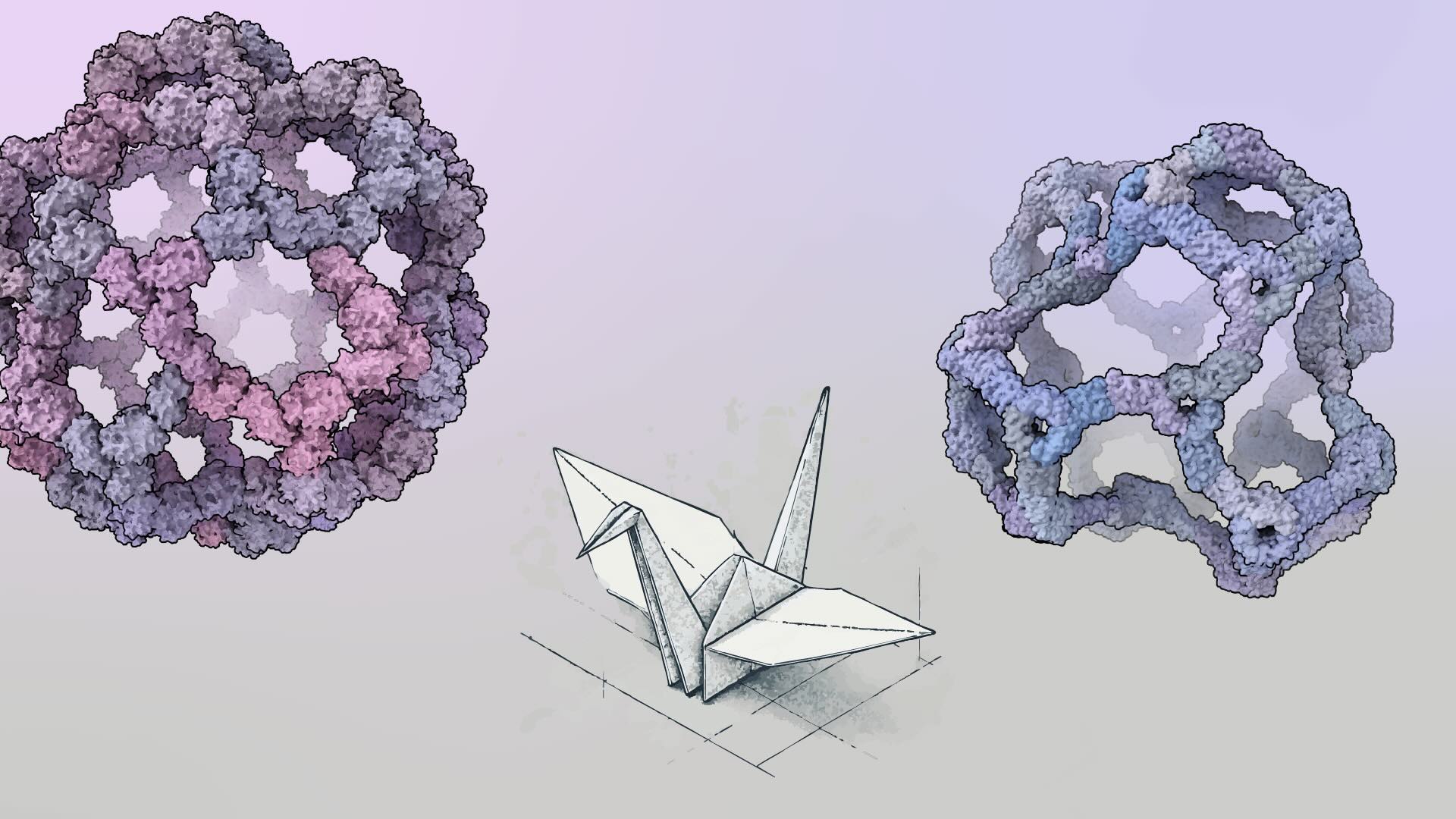
Building bigger nanocages with less symmetry
New protein assemblies with hundreds of subunits dwarf common viral capsids.
-
Introducing RFpeptides
Generative AI based on RFdiffusion designs cyclic peptides with atomic accuracy.
-
Webinar: Protein Design Then & Now (1988-2024)
Computational protein design, once considered impossible, has blossomed into a transformative research field with implications for medicine, sustainability, and beyond. In a recent webinar hosted by David Baker, three pioneers in the field—Bill DeGrado, Steve Mayo, and Brian Kuhlman—shared their insights on the origins, evolution, and future of protein design.
-
David Baker wins Nobel Prize for protein design
“Baker has learned how to master life’s building blocks and create entirely new proteins.”