By harnessing the potential of proteins, we are pioneering a new wave of medicines and materials with revolutionary capabilities.
Research Areas

Protein Structure Prediction
We develop cutting-edge tools, like RoseTTAFold, that predict the three-dimensional structures of proteins from sequence data with remarkable speed and accuracy. These tools are available for free public use, making advanced protein modeling accessible to researchers worldwide.
Protein & Small Molecule Binding
We create methods for generating proteins that bind to targets of medical interest. This research deepens our understanding of the molecular interactions that drive cellular processes, paving the way for new therapeutic strategies.
Self-Assembling Nanomaterials
We design self-assembling nanostructures that have applications as vaccines, drug delivery systems, and more. Our first approved medicine — a COVID-19 vaccine — leverages this technology, showcasing the potential of computationally designed protein nanoparticles.
New Protein Scaffolds
In the past, almost all protein design efforts involved modifying proteins from nature. We are developing methods for designing a wide range of exceptionally stable protein folds from scratch with tunable features for specific applications.
Enzyme Design
Enzymes catalyze chemical reactions that are essential for life. We are developing methods to design new enzymes for chemical transformations not found in nature. This research helps reveal more about the chemical foundations of life.
Protein Structure Determination
Our tools for solving protein structures with limited experimental data are used by researchers around the globe, accelerating the discovery of new protein functions and interactions.
Applications
From detecting fentanyl to blocking the virus that causes COVID-19, our scientists are pushing the limits of what proteins can do to help meet the challenges of the modern world.

Ultrapotent Vaccines
We played a key role in developing SKYCovione, the first medicine ever created using computational protein design. This COVID-19 vaccine, featuring 60 copies of the SARS-CoV-2 Spike protein receptor binding domain, has been approved in multiple countries and received an Emergency Use Listing from the World Health Organization. Our work also includes the creation of RSV-targeting nanoparticle vaccines, which led to the launch of the spinout Icosavax, later acquired by AstraZeneca. We are currently developing broad-spectrum vaccines against influenza, coronaviruses, and more, aiming to provide robust protection against future viral threats.
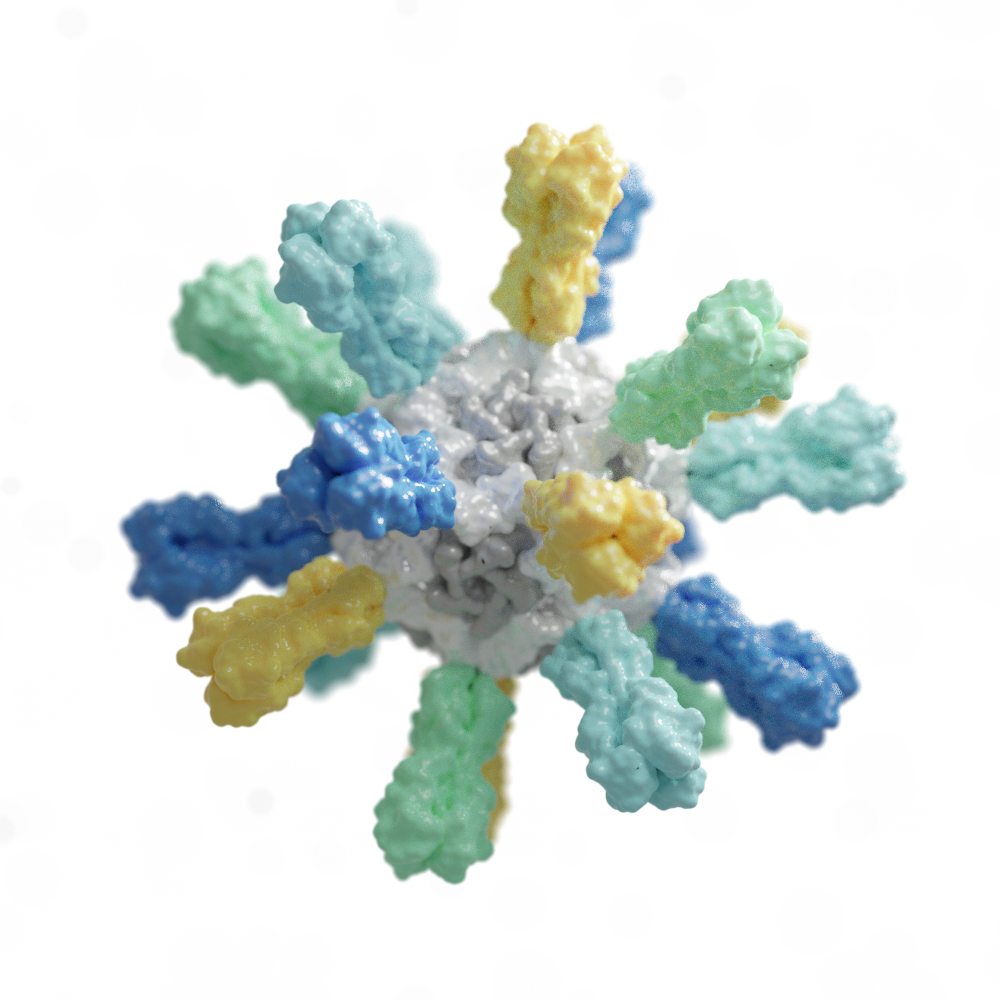
Rapid Antivirals
Since 2011, we have been designing proteins that inhibit viruses such as influenza. These computer-generated antivirals are not only durable and effective in animal models but also serve as enhanced reagents in low-cost diagnostic tests, outperforming traditional antibody-based systems.
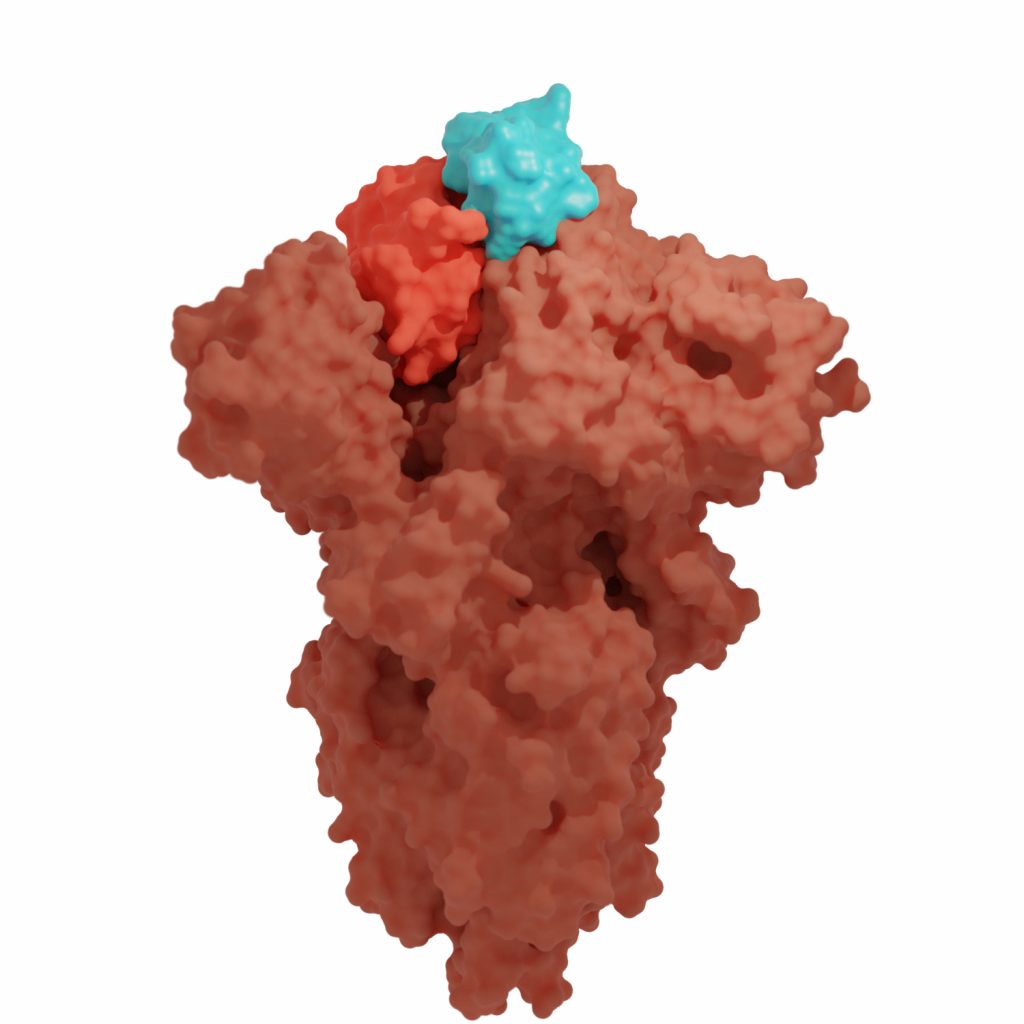
Cancer Immunotherapy
We’re creating synthetic proteins that function as powerful cancer immunotherapies without triggering off-target effects. These compact proteins have shown promise in shrinking tumors in animal models, offering a potential pathway to safer cancer treatments.
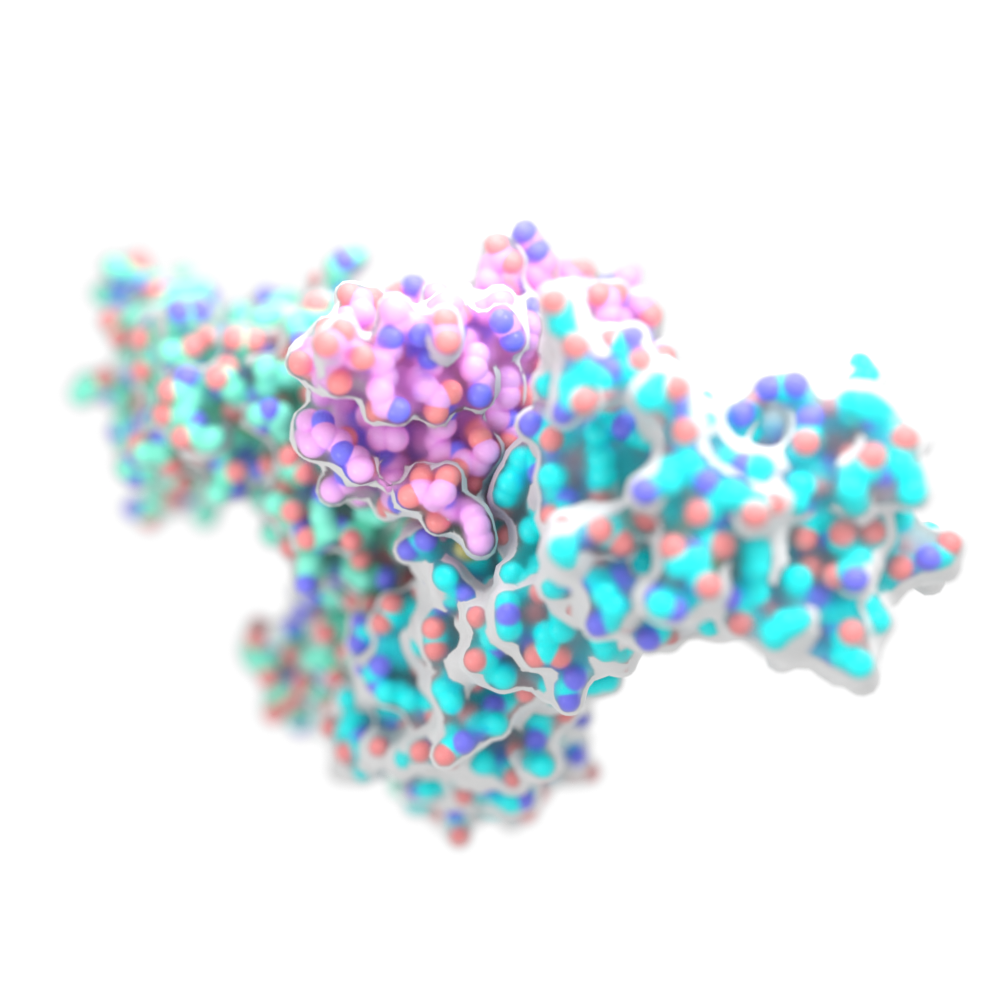
Celiac Disease
Building on an iGEM-winning student project from the Baker Lab, we developed Kuma030, an enzyme engineered to break down gluten in the stomach, mitigating the symptoms of celiac disease. This enzyme was further developed by our spinout company PvP Biologics, which has been acquired by Takeda.
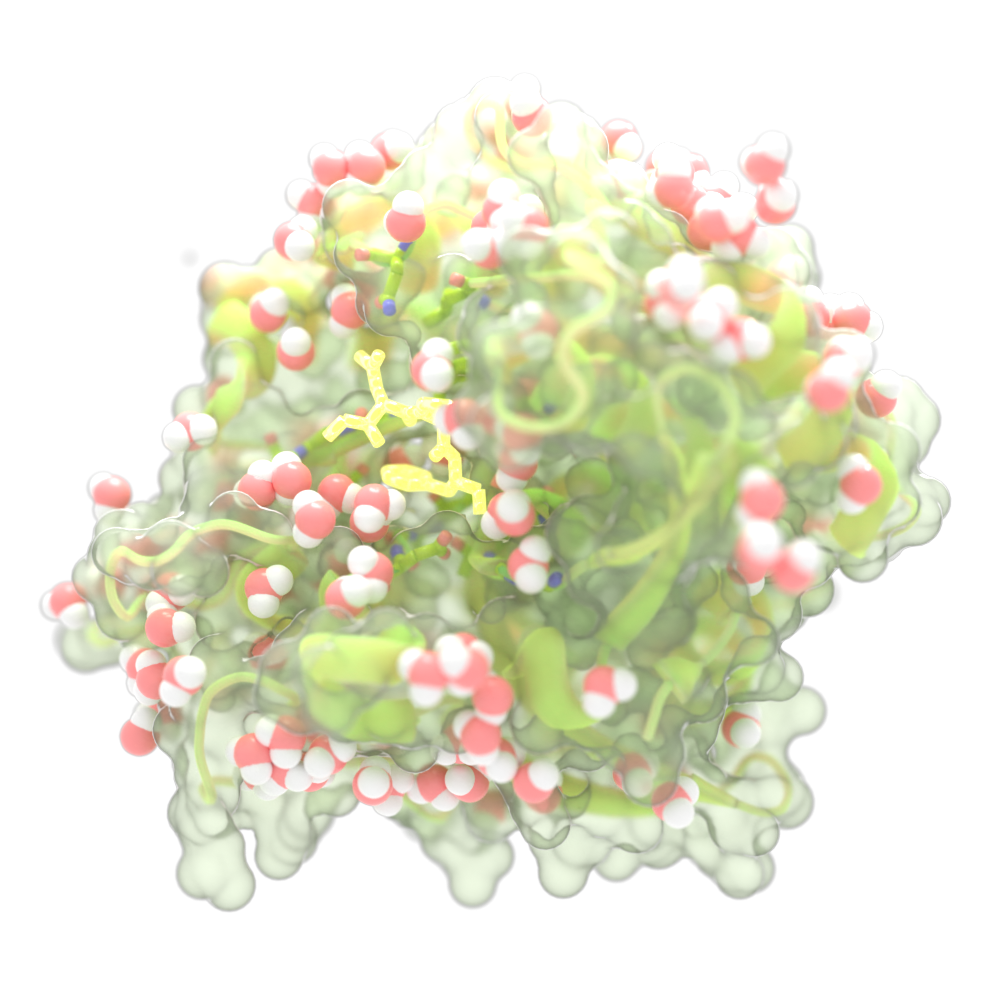
New Nanomaterials
Inspired by natural materials like silk and bone, our researchers are creating new biomaterials at the nanoscale. These advanced materials have the potential to replace traditional, polluting substances with sustainable alternatives, opening the door to groundbreaking new applications.

Research alone isn’t enough to change the world — that’s why we launch new companies.
