References
- Koday MT, Nelson J, Chevalier A, Koday M, Kalinoski H, Stewart L, Carter L, Nieusma T, Lee PS, Ward AB, Wilson IA, Dagley A, Smee DF, Baker D, Fuller DH: A Computationally Designed Hemagglutinin Stem-Binding Protein Provides In Vivo Protection from Influenza Independent of a Host Immune Response. PLoS pathogens 2016, 12(2):e1005409. PMC#
- Holstein CA, Chevalier A, Bennett S, Anderson CE, Keniston K, Olsen C, Li B, Bales B, Moore DR, Fu E, Baker D, Yager P: Immobilizing affinity proteins to nitrocellulose: a toolbox for paper-based assay developers. Anal Bioanal Chem 2016, 408(5):1335-1346. PMC#
- Silva DA, Yu S, Ulge UY, Spangler JB, Jude KM, Labão-Almeida C, Ali LR, Quijano-Rubio A, Ruterbusch M, Leung I, Biary T, Crowley SJ, Marcos E, Walkey CD, Weitzner BD, Pardo-Avila F, Castellanos J, Carter L, Stewart L, Riddell SR, Pepper M, Bernardes GJL, Dougan M, Garcia CK, Baker D: De novo design of potent and selective mimics of IL-2 and IL-15. Nature 2019, 565(7738):186-191. PMC#30626941
- Mazor R, Eberle JA, Hu X, Vassall AN, Onda M, Beers R, Lee EC, Kreitman RJ, Lee B, Baker D, King C, Hassan R, Benhar I, Pastan I: Recombinant immunotoxin for cancer treatment with low immunogenicity by identification and silencing of human T-cell epitopes. Proceedings of the National Academy of Sciences of the United States of America 2014, 111(23):8571-8576. PMC#4060717
- Wolf C, Siegel JB, Tinberg C, Camarca A, Gianfrani C, Paski S, Guan R, Montelione G, Baker D, Pultz IS: Engineering of Kuma030: A Gliadin Peptidase That Rapidly Degrades Immunogenic Gliadin Peptides in Gastric Conditions. Journal of the American Chemical Society 2015, 137(40):13106-13113. PMC#
- Gordon SR, Stanley EJ, Wolf S, Toland A, Wu SJ, Hadidi D, Mills JH, Baker D, Pultz IS, Siegel JB: Computational design of an alpha-gliadin peptidase. Journal of the American Chemical Society 2012, 134(50):20513-20520. PMC#3526107
- Ofek G, Guenaga FJ, Schief WR, Skinner J, Baker D, Wyatt R, Kwong PD: Elicitation of structure-specific antibodies by epitope scaffolds. Proceedings of the National Academy of Sciences of the United States of America 2010, 107(42):17880-17887. PMC#PMC2964213
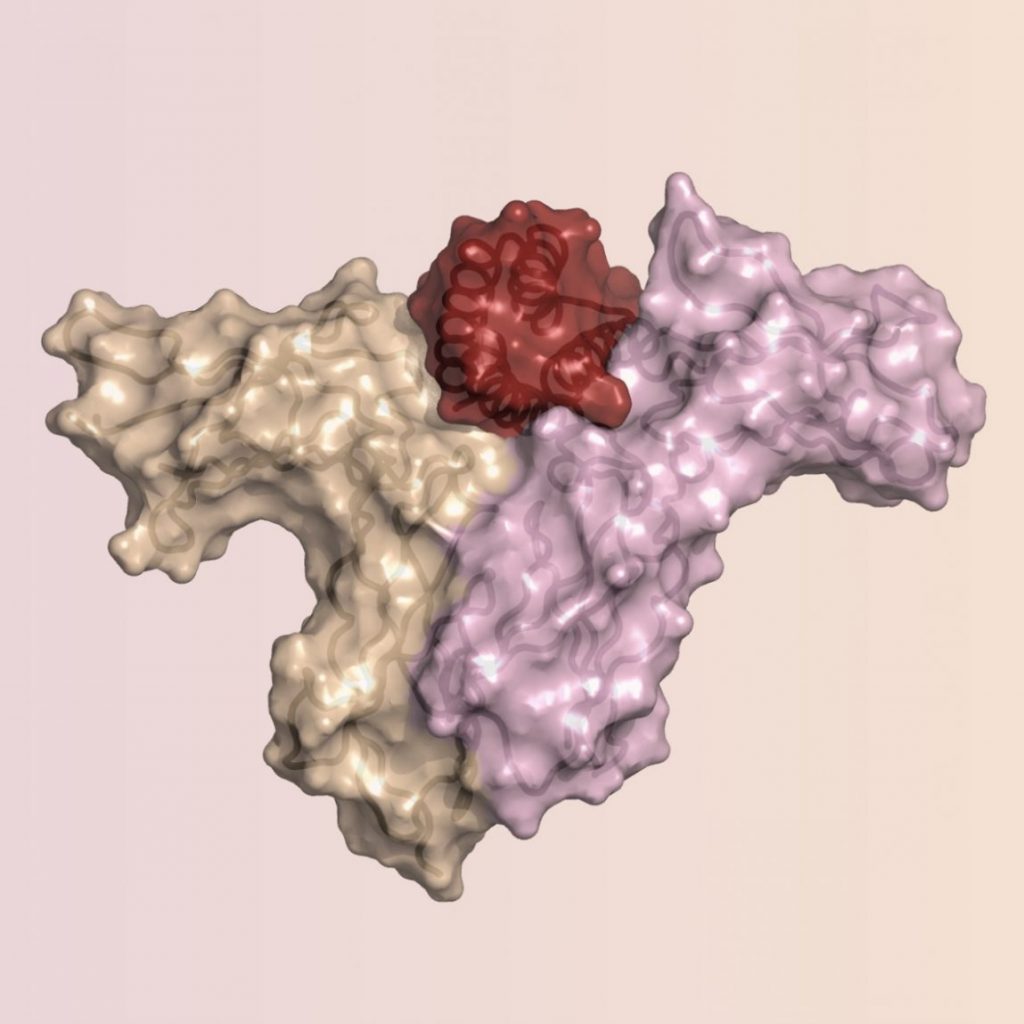
- Correia BE, Bates JT, Loomis RJ, Baneyx G, Carrico C, Jardine JG, Rupert P, Correnti C, Kalyuzhniy O, Vittal V, Connell MJ, Stevens E, Schroeter A, Chen M, Macpherson S, Serra AM, Adachi Y, Holmes MA, Li Y, Klevit RE et al: Proof of principle for epitope-focused vaccine design. Nature 2014, 507(7491):201-206. PMC#PMC4260937
- Correia BE, Ban YE, Holmes MA, Xu H, Ellingson K, Kraft Z, Carrico C, Boni E, Sather DN, Zenobia C, Burke KY, Bradley-Hewitt T, Bruhn-Johannsen JF, Kalyuzhniy O, Baker D, Strong RK, Stamatatos L, Schief WR: Computational design of epitope-scaffolds allows induction of antibodies specific for a poorly immunogenic HIV vaccine epitope. Structure 2010, 18(9):1116-1126. PMC#
- Griss R, Schena A, Reymond L, Patiny L, Werner D, Tinberg CE, Baker D, Johnsson K: Bioluminescent sensor proteins for point-of-care therapeutic drug monitoring. Nature chemical biology 2014, 10(7):598-603. PMC#
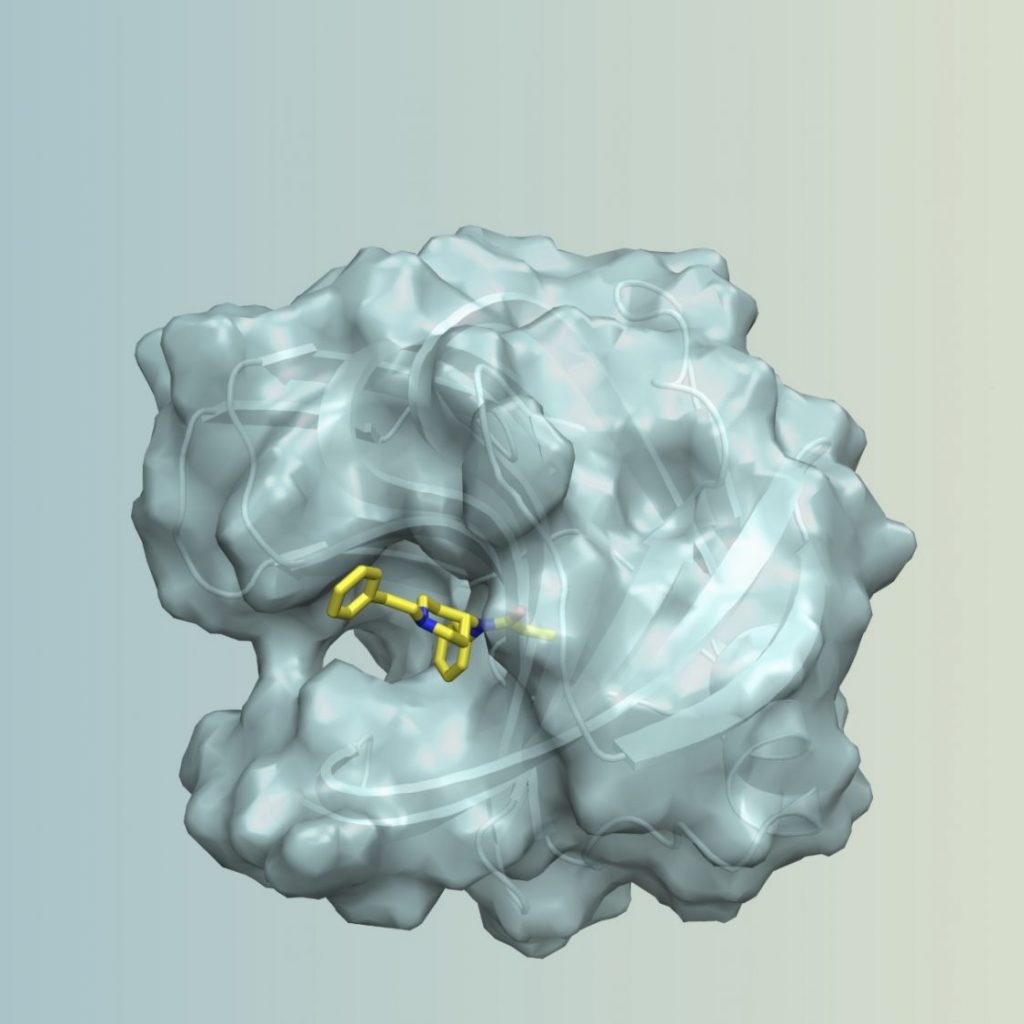
- Tinberg CE, Khare SD, Dou J, Doyle L, Nelson JW, Schena A, Jankowski W, Kalodimos CG, Johnsson K, Stoddard BL, Baker D: Computational design of ligand-binding proteins with high affinity and selectivity. Nature 2013, 501(7466):212-216. PMC#3898436
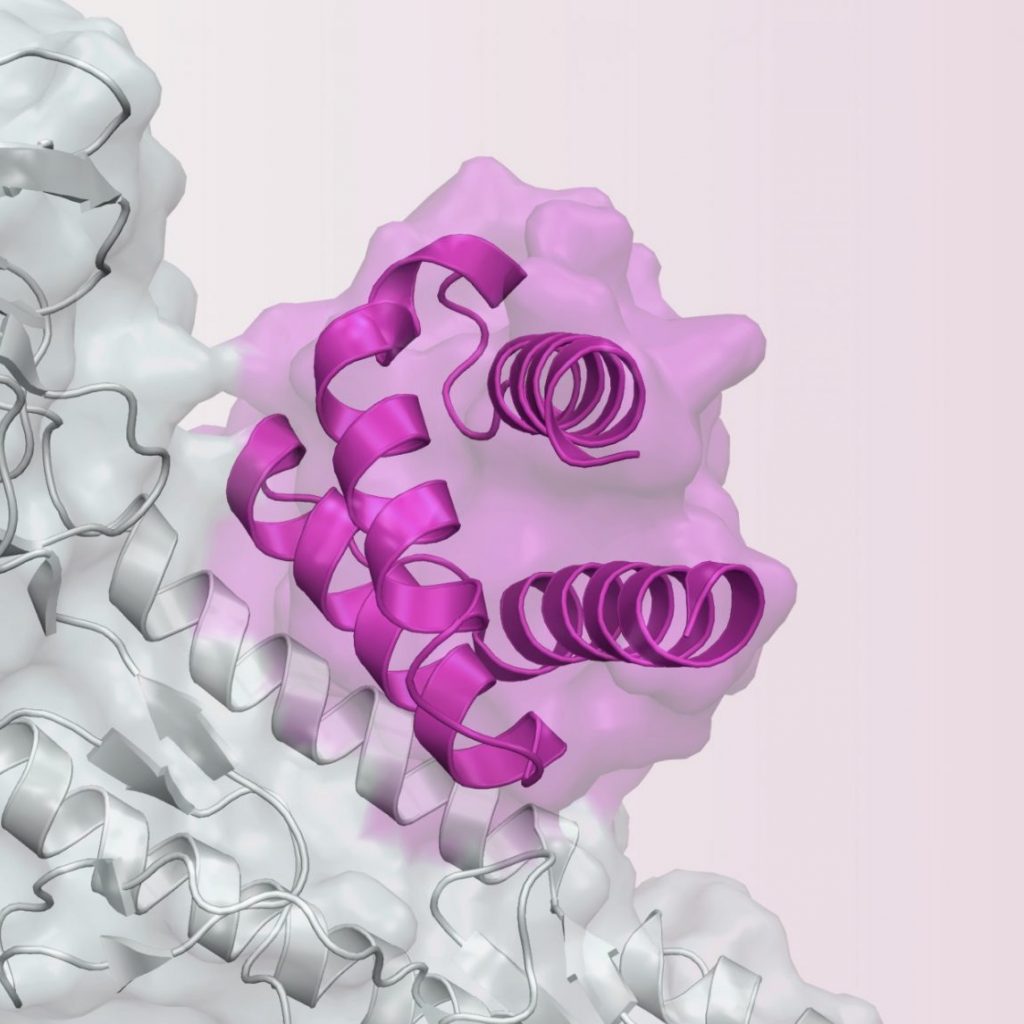
- Taylor ND, Garruss AS, Moretti R, Chan S, Arbing MA, Cascio D, Rogers JK, Isaacs FJ, Kosuri S, Baker D, Fields S, Church GM, Raman S: Engineering an allosteric transcription factor to respond to new ligands. Nature methods 2016, 13(2):177-183. PMC#
- Feng J, Jester BW, Tinberg CE, Mandell DJ, Antunes MS, Chari R, Morey KJ, Rios X, Medford JI, Church GM, Fields S, Baker D: A general strategy to construct small molecule biosensors in eukaryotes. eLife 2015, 4. PMC#PMC4739774
- Fleishman SJ, Whitehead TA, Ekiert DC, Dreyfus C, Corn JE, Strauch EM, Wilson IA, Baker D: Computational design of proteins targeting the conserved stem region of influenza hemagglutinin. Science 2011, 332(6031):816-821. PMC#3164876
- King C, Garza EN, Mazor R, Linehan JL, Pastan I, Pepper M, Baker D: Removing T-cell epitopes with computational protein design. Proceedings of the National Academy of Sciences of the United States of America 2014, 111(23):8577-8582. PMC#4060723
- Bick MJ, Greisen PJ, Morey KJ, Antunes MS, La D, Sankaran B, Reymond L, Johnsson K, Medford JI, Baker D, Cravatt, BF: Computational design of environmental sensors for the potent opioid fentanyl. eLife 2017, 6. PMC#5655540
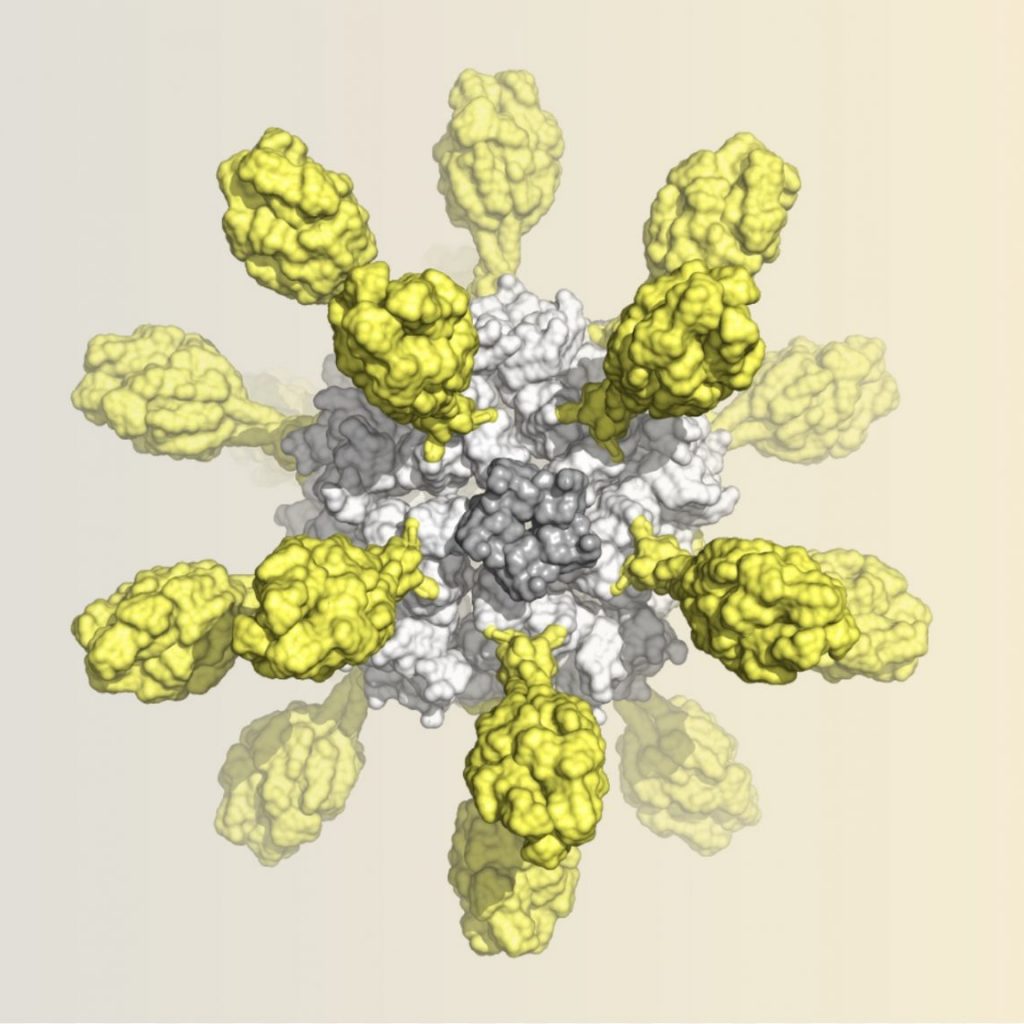
- Walls AC, Fiala B, Schäfer S, […] , Helen Y. Chu HY, Lee KK, Fuller DH, Baric RS, Kellam P, Carter L, Pepper M, Sheahan TP, Veesler D, King NP: Elicitation of potent neutralizing antibody responses by designed protein nanoparticle vaccines for SARS-CoV-2. Cell 2020, PMC#7604136
- Cao L, Goreshnik I, Coventry B, Case, JB, Miller L […], Diamond, MS; Veesler D, Baker D: De novo design of picomolar SARS-CoV-2 miniprotein inhibitors. Science 2020, PMC#7857403
- Quijano-Rubio A, Yeh H, Park J, Lee H, Langan RA, Boyken SE, Lajoie ML, Cao L, Chow CM, Miranda MC, Wi J, Hong HJ, Stewart L, Oh B-H Baker D: De novo design of modular and tunable allosteric biosensors. Nature, 2021. PMC#7386493