Our Response to COVID-19
“This outbreak has illustrated that it’s all hands on deck, and all of us together against the bugs. We are working with our collaborators at UW, the NIH, and the Bill & Melinda Gates Foundation to help create a safe and effective vaccine for not only SARS-CoV-2 but other coronaviruses as well.” – Neil King, head of vaccine design



Potent Vaccines
We delivered the world’s first medicine developed using computational protein design. This approach uses software to engineer vaccines with enough precision to place each atoms exactly where desired.
A vaccine for COVID-19 that uses our self-assembling nanoparticle technology has been authorized in the United Kingdom and South Korea. The vaccine, now called SKYCovione (SKYCovion in the UK), has also been granted an Emergency Use Listing by the World Health Organization.
The vaccine’s clinical development is being led by our collaborators at SK Bioscience. The University of Washington is licensing the vaccine technology royalty-free for the duration of the pandemic.
The Dream Vaccine | Science
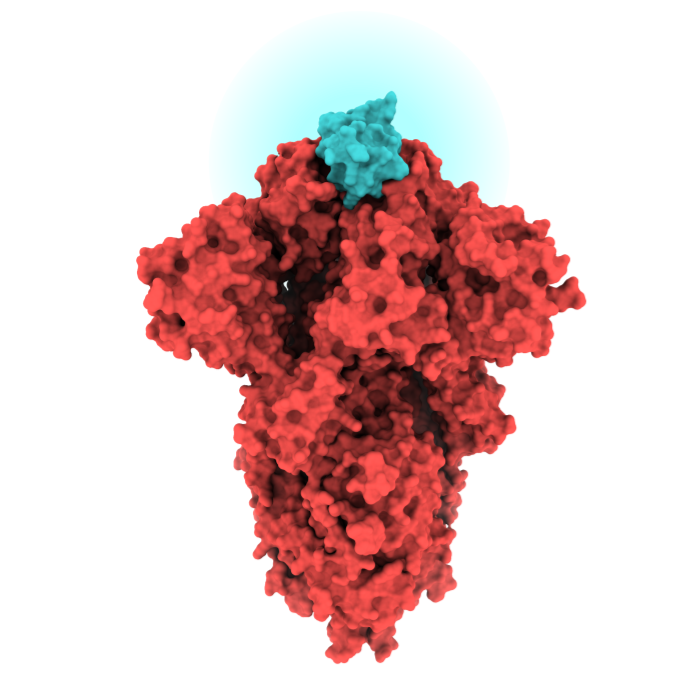
Rapid Antivirals
In less than a month, our researchers designed over two million antiviral proteins targeting the novel coronavirus.
Over 120,000 of the most promising candidates were tested in the lab. Some neutralize all strains of the novel coronavirus with activities rivaling the best-known antibodies. We are working with collaborators to advance these candidate antivirals into clinical testing.
De novo design of picomolar SARS-CoV-2 miniprotein inhibitors | Science
Antibodies Good. Machine-Made Molecules Better? | New York Times
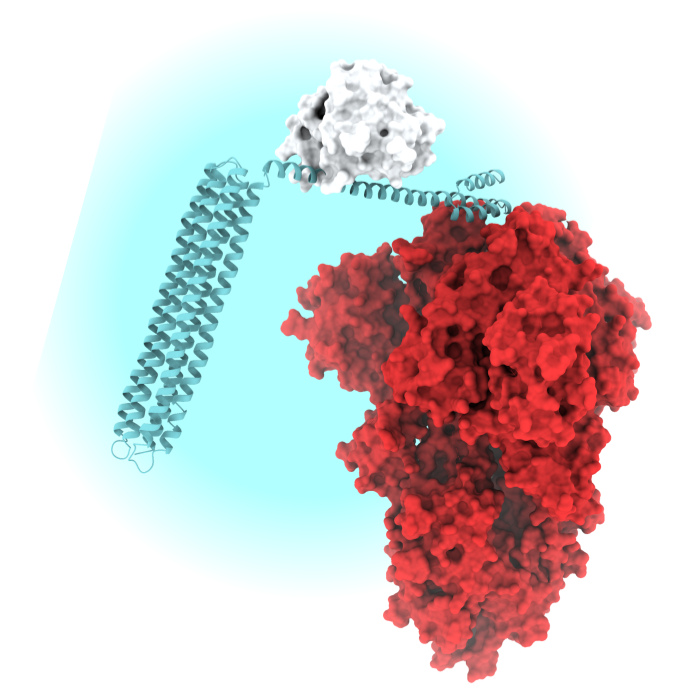
Innovative Diagnostics
Our scientists have created new ways to detect SARS-CoV-2 in blood, as well as protective antibodies against the virus.
Diagnostic tests are essential for detecting coronavirus infection and immunity. These biosensors are currently being optimized for use without laboratory instruments or refrigeration.
De novo design of modular and tunable protein biosensors | Nature
Support Tomorrow’s Innovations
Our ability to respond quickly to the COVID-19 crisis was due in part to early supporters, leadership funding from the Bill & Melinda Gates Foundation, and transformative investment from The Audacious Project. Visionary philanthropists can still help provide the resources needed to continue advancing our pandemic preparedness and response research.
If you would like to make a gift to support the Institute for Protein Design, please contact Katherine Cardinal, Associate Vice President for Development at UW Medicine, at 206.650.4503 or cardinal@uw.edu.

