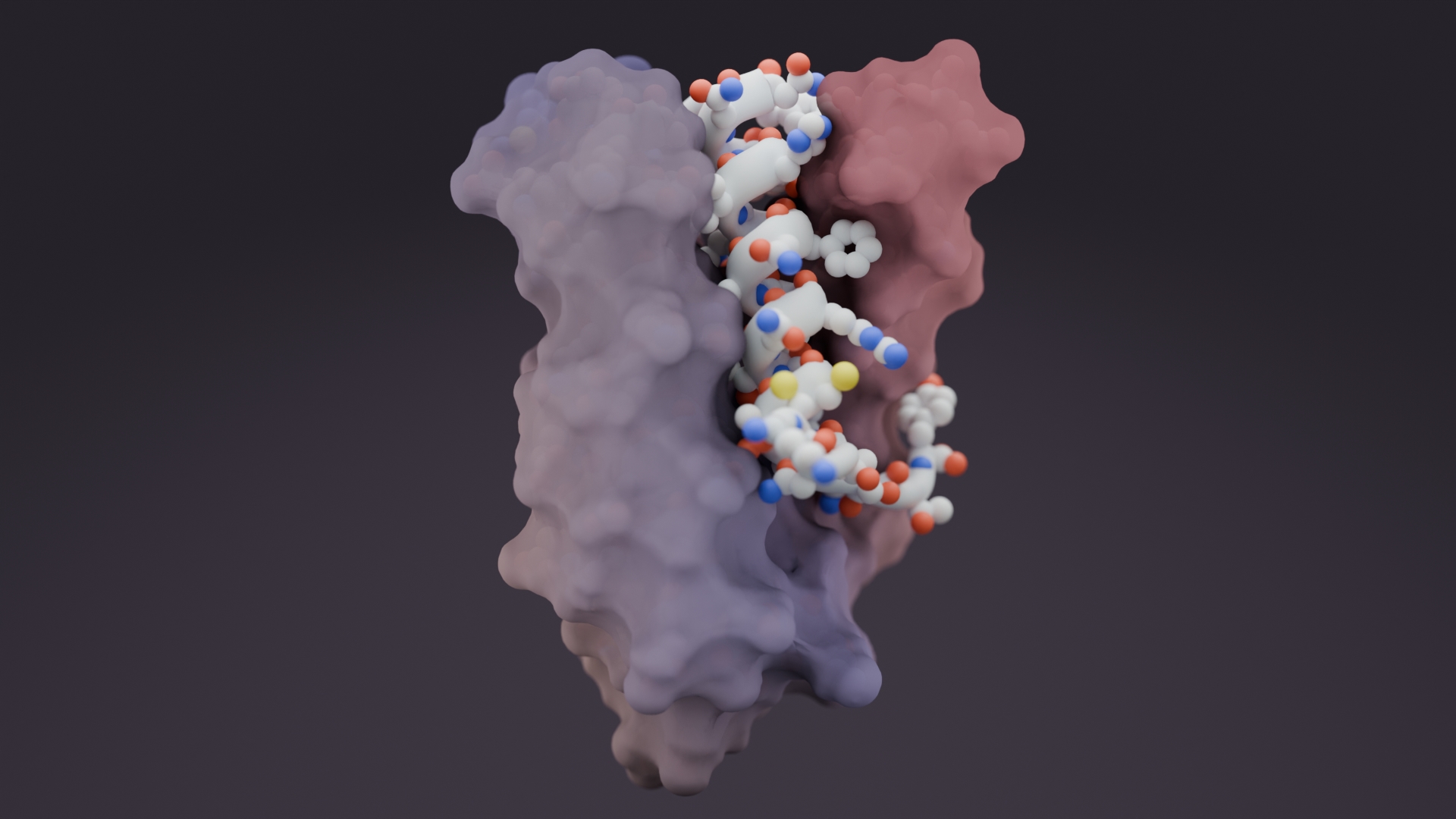
Researchers in the Baker lab at the Institute for Protein Design, working in collaboration with the Joint Genome Institute, published in Science the solved folds and structures for hundreds of protein families. This “big data” approach to large scale protein structure determination was made possible by a team effort that analyzed billions of gene sequences read out from soil, ocean, and air samples collected around the globe.
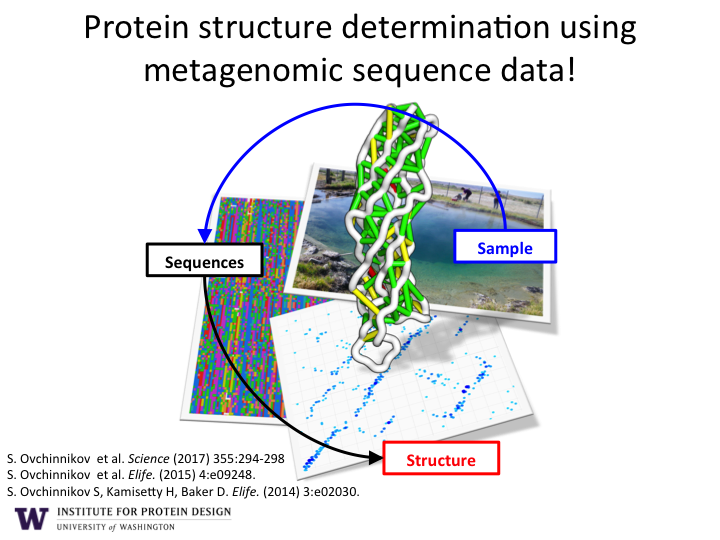
The research has been recognized by numerous opinion leaders and media outlets as an unprecedented breakthrough for protein structure prediction. See articles in The Atlantic, The Economist , Science, GeekWire, and GEN.
How does it work?
As illustrated in Figure 1, the sequencing of DNA from environmental samples produces billions of new protein amino acid sequences. Computer algorithms are used to align the sequences according to their evolutionary history. This allows the discovery of pairs of amino acids that co-evolve. If a change occurs in one amino acid, then a compensatory change is typically observed in another amino acid in the sequence. Co-evolving pairs of amino acids are almost always in close proximity to each other (green and yellow lines) within in the final 3D structure of the protein structure (white backbone).
Why is it important?
With this approach, the team produced reliable models for 622 protein families, and discovered more than 100 new protein folds. In addition to resolving the folding structure of a protein, as shown in Figure 2 co-evolution data can also provide data on the dynamic nature of protein structure including transient contacts, protein-protein contacts, and contacts with ligands. Over time, as more environmental DNA sequence data becomes available, we expect to greatly increase our understanding of protein structure, assembly, and function. In turn, we expect this information to enable the design of new proteins with functions.
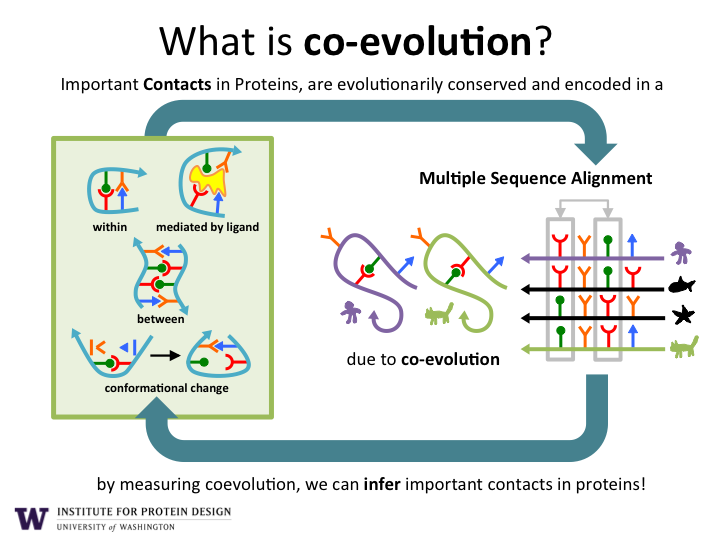
Sharing data.
The Institute for Protein Design believes in sharing its insights with the rest of the world and we have made publicly available the database of protein structures resolved by these methods.