Category: Publication
-

Introducing RFdiffusion2
A new deep-learning model developed by scientists at the IPD and MIT is redefining what’s possible in enzyme design.
-

Introducing LigandMPNN
ProteinMPNN revolutionized protein sequence design. LigandMPNN opens the door to programming protein–ligand interfaces of all kinds.
-

Neutralizing deadly snake toxins
New AI-generated proteins that protect animals in the lab promise safer, more accessible antivenoms.
-
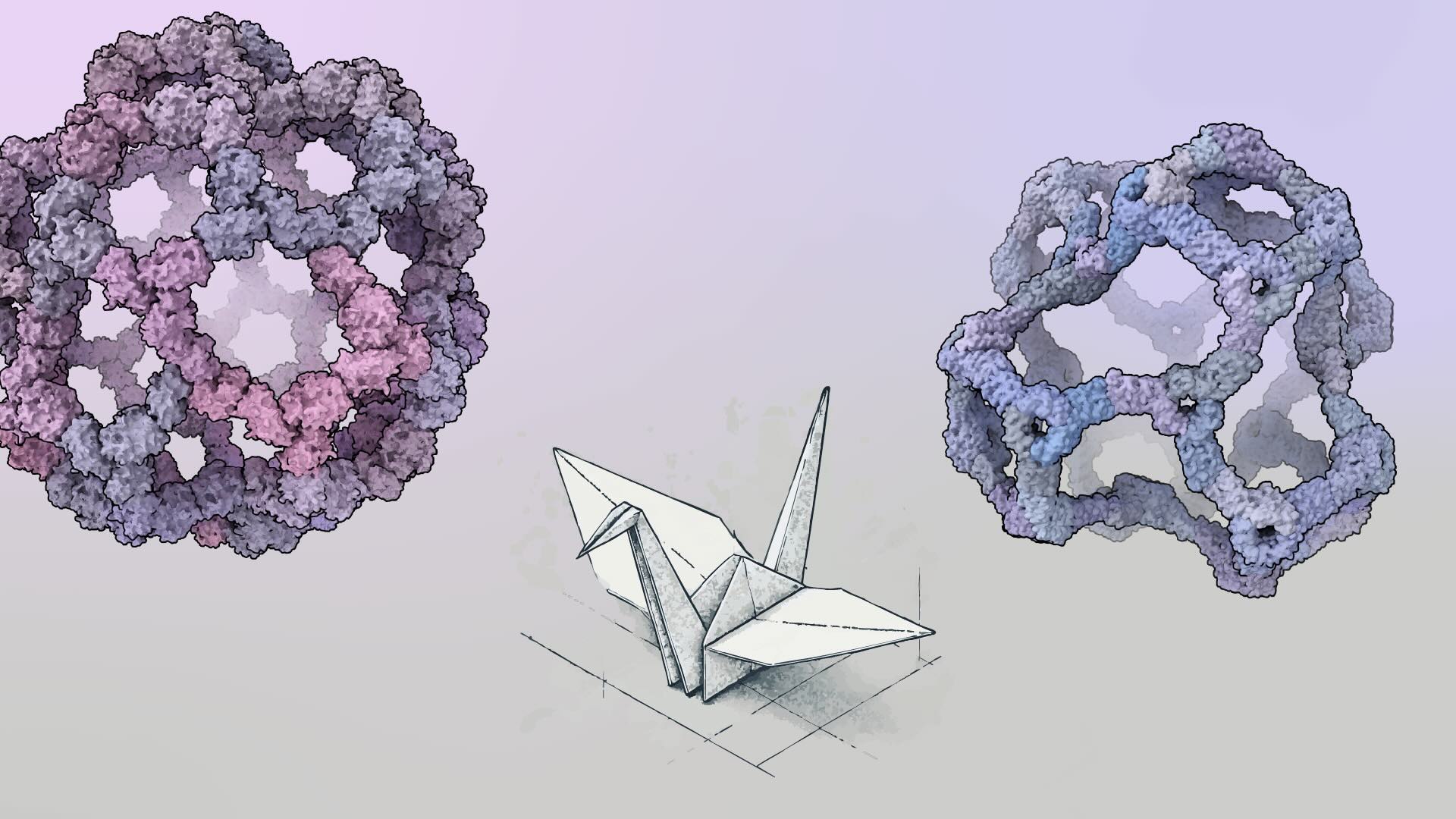
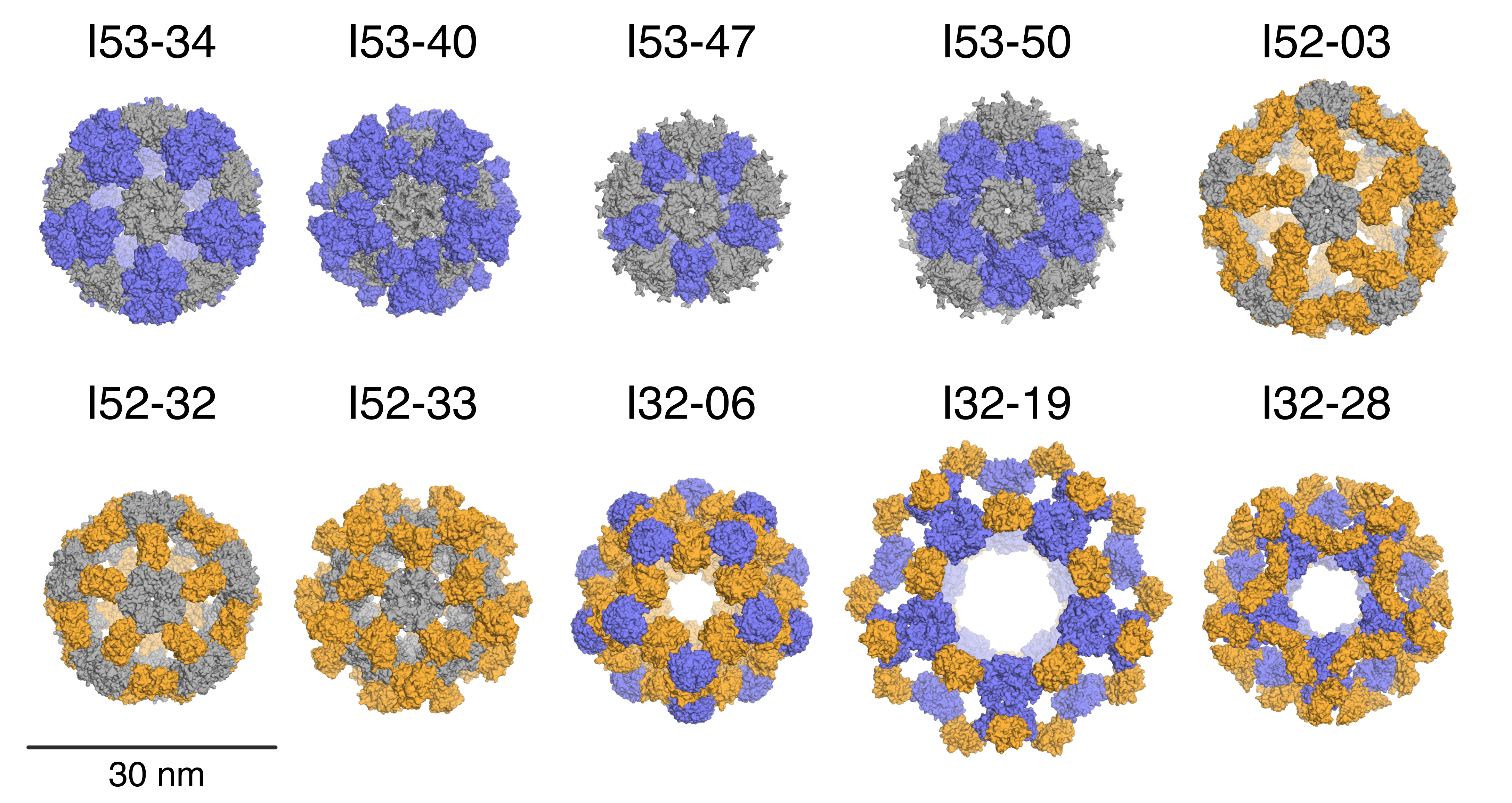
Building bigger nanocages with less symmetry
New protein assemblies with hundreds of subunits dwarf common viral capsids.
-

Introducing RFpeptides
Generative AI based on RFdiffusion designs cyclic peptides with atomic accuracy.
-

Designing synthetic smell receptors
“When we started with this idea a few years ago, many people thought it was impossible.”
-

AI generates proteins with exceptional binding strengths
This advance could allow scientists to create cheaper alternatives to antibodies for disease detection and treatment. This week we report in Nature an AI-enabled advance in biotechnology with implications for drug development, disease detection, and environmental monitoring. Using a combination of traditional and deep learning based molecular design approaches, we’ve created proteins that bind with…
-

A new path to carbon storage
Our research shows that custom proteins can drive the growth of limestone-like minerals, a breakthrough that may one day help remove excess carbon from the environment.
-

Introducing All-Atom versions of RoseTTAFold and RFdiffusion
Recent deep-learning breakthroughs allow us to look beyond just amino acids, expanding the types of proteins that can be modeled and designed using RoseTTAFold and RFdiffusion.
-

Nature: “AI tools are designing entirely new proteins that could transform medicine”
RFdiffusion was published in Nature this week. The journal reports on how it and other generative algorithms are being used to create custom biomolecules in seconds.
-

Top-down design of protein architectures with reinforcement learning
Today we report in Science [PDF] the successful application of reinforcement learning to a challenge in protein design. This research is a milestone in the use of artificial intelligence for science, and the potential applications are vast, from developing more effective cancer treatments to new biodegradable textiles. A team led by scientists in the Baker…
-

Design of binders for disordered targets
Today we report in Nature the design of proteins that recognize and bind to the so-called “intrinsically disordered regions” of proteins and peptides. The body produces such disordered molecules naturally, but many have been linked to health disorders, including myeloma and other cancers. “Disordered proteins play important roles in biology. By designing new proteins that…
-
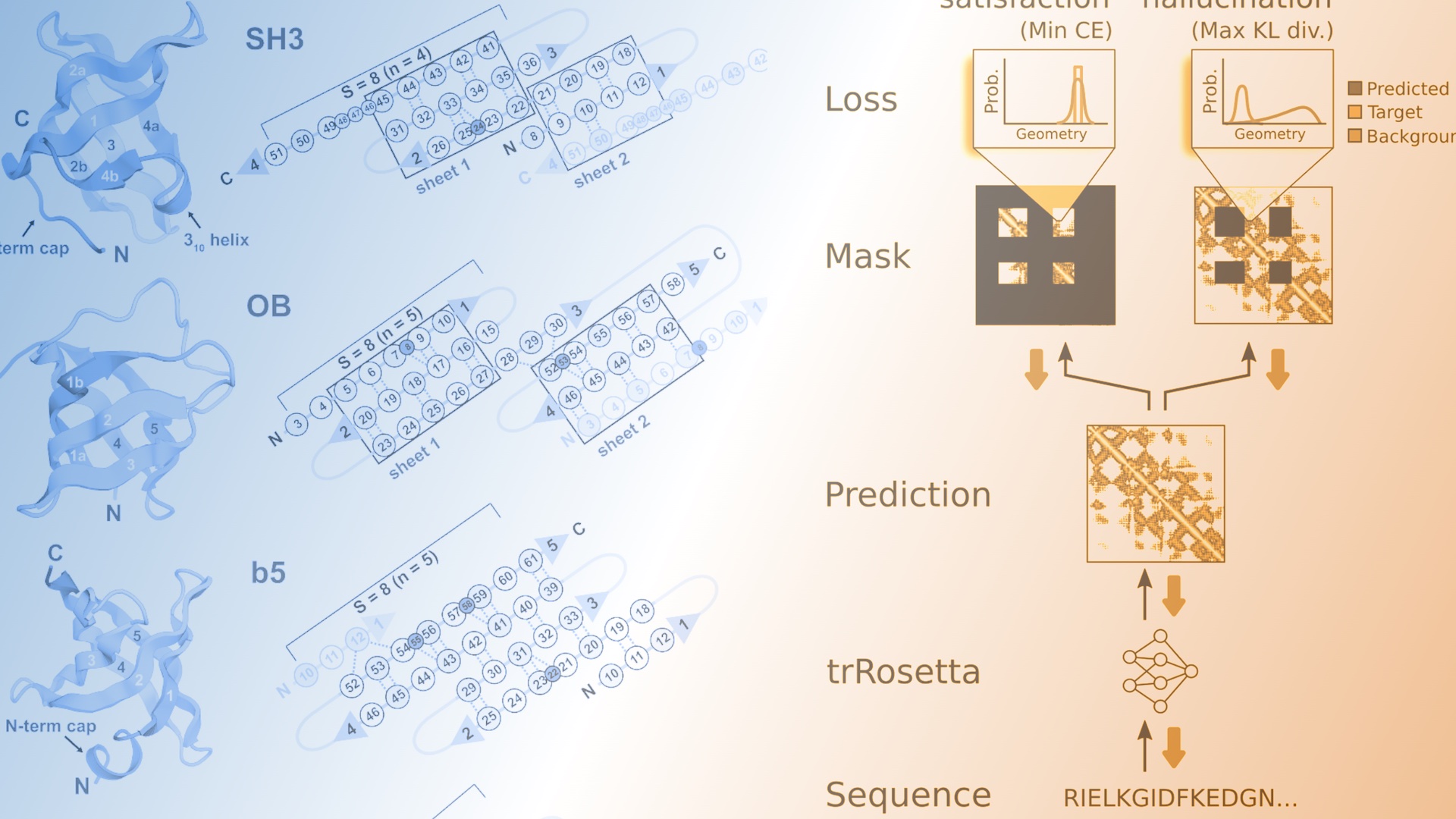
De novo design of small beta barrel proteins
The de novo design of small proteins with beta-barrel topologies has been a challenge for computational design due to the complexity inherent in these folds. In a new study appearing in PNAS, a team led by Baker Lab research scientist David E. Kim describes the successful design and characterization of four different classes of small…
-

Degreaser: Improving protein secretion
King Lab postdoctoral scholars John Wang and Alena Khmelinskaia have developed a new tool for identifying and removing hidden transmembrane sequences that hinder protein secretion.
-
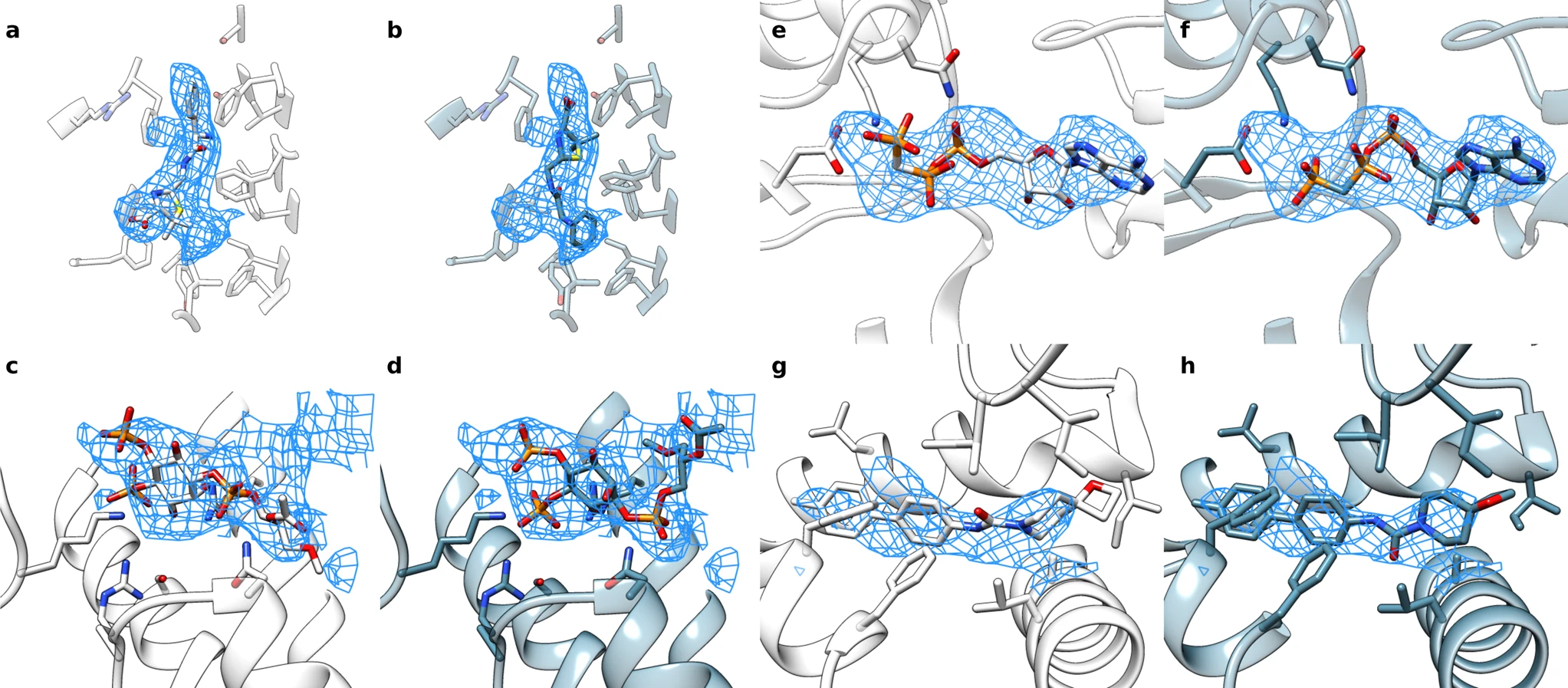
EMERALD: Automatically locate ligands in cyroEM maps
Under ideal conditions, cryo-electron microscopy can be used to determine protein structures at near-atomic resolution. But conditions are not always perfect. To help researchers make use of medium-resolution cryo-electron density maps, scientists in the DiMaio Lab have developed EMERALD, which is a new software tool that can accurately and automatically produce deposition-ready small molecule models…
-

Machine learning generates custom enzymes
Today we report in Nature the computational design of highly efficient enzymes unlike any found in nature. Laboratory testing confirms that the new light-emitting enzymes can recognize specific chemical substrates and catalyze the emission of photons very efficiently.
-

A diffusion model for protein design
Update (July 2023): Our manuscript on the development of RFdiffusion has been published in Nature. A team led by scientists from the Baker Lab has created a powerful new way of designing proteins that combines structure prediction networks and generative diffusion models. The team demonstrated extremely high computational success using the new method and experimentally…
-

ProteinMPNN excels at creating new proteins
Over the past two years, machine learning has revolutionized protein structure prediction. Now, three papers in Science describe a similar revolution in protein design. In the new papers, scientists in the Baker lab show that machine learning can be used to create proteins much more accurately and quickly than previously possible. This could lead to…
-
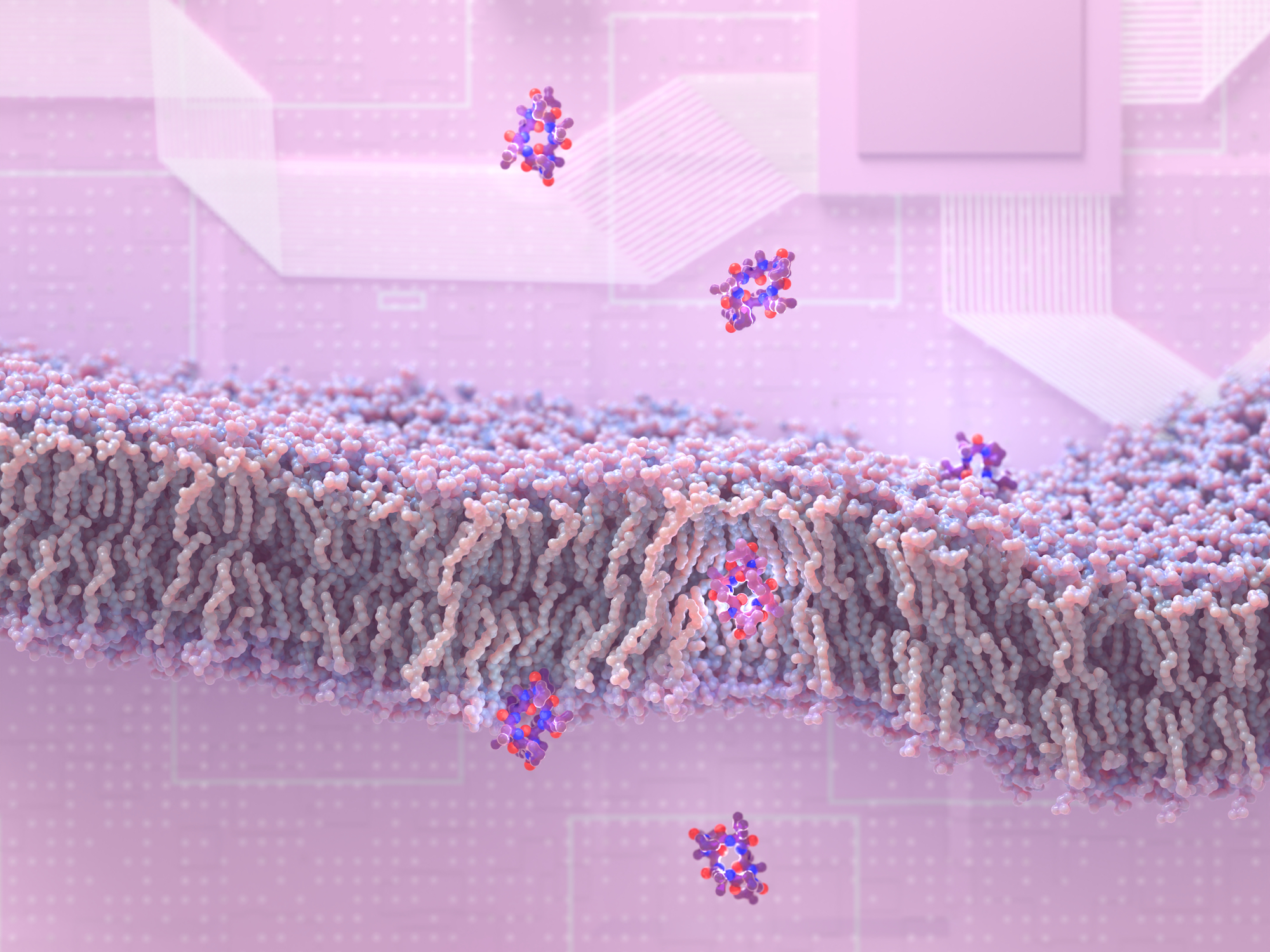
Design of membrane-traversing peptides leads to new spinout
Researchers at the Institute for Protein Design have discovered how to create peptides that slip through membranes and enter cells. This drug design breakthrough may lead to new medications for a wide variety of health disorders, including cancer, infection, and inflammation. This research appears in the journal Cell [PDF]. “This new ability to design membrane-permeable…
-
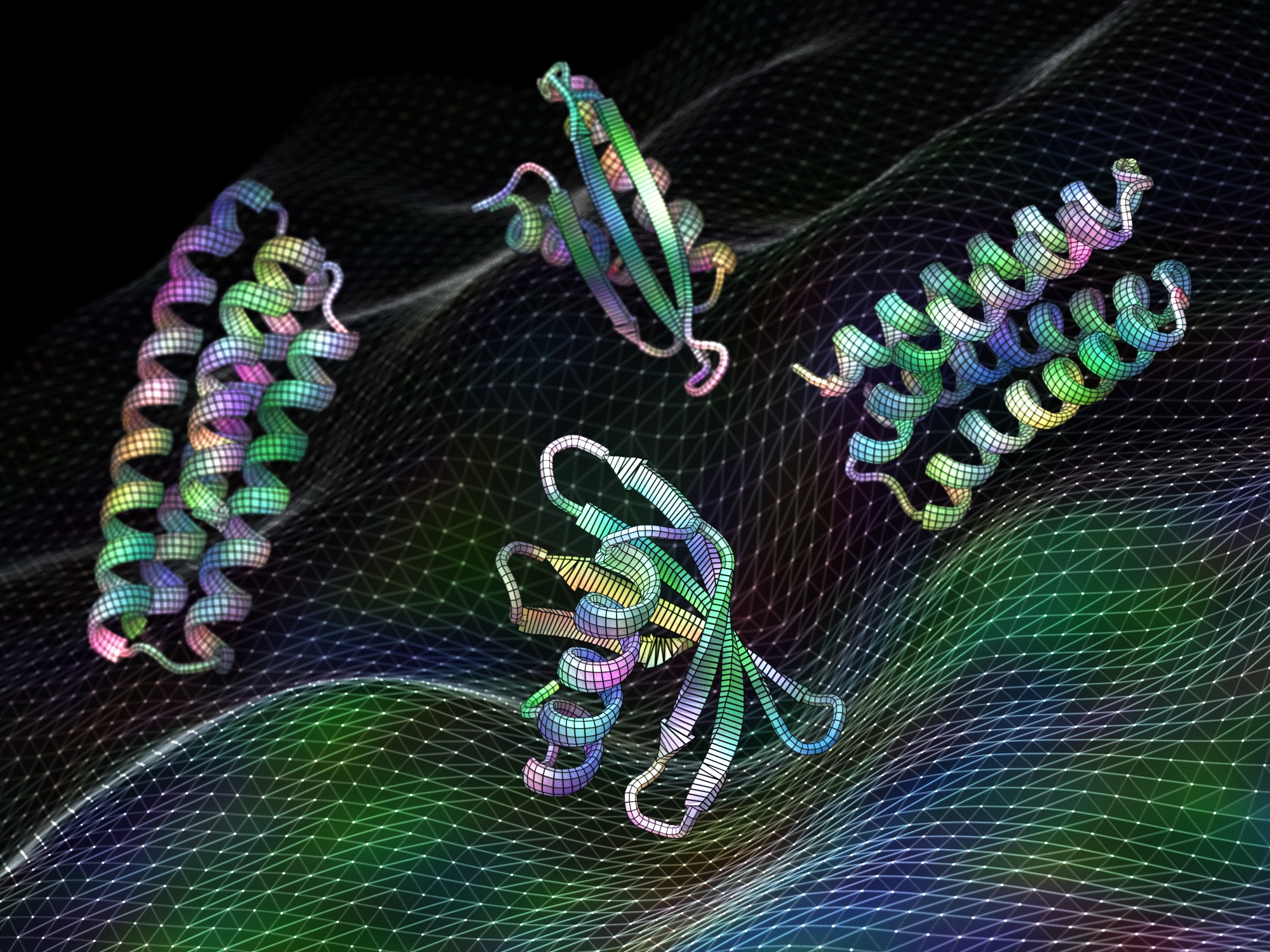
Training A.I. to generate medicines and vaccines
Today we report in Science [PDF] the development of artificial intelligence software that can create proteins that may be useful as vaccines, cancer treatments, or even tools for pulling carbon pollution out of the air. This project was led by Jue Wang, Doug Tischer, and Joseph L. Watson, who are postdoctoral scholars at UW Medicine,…
-
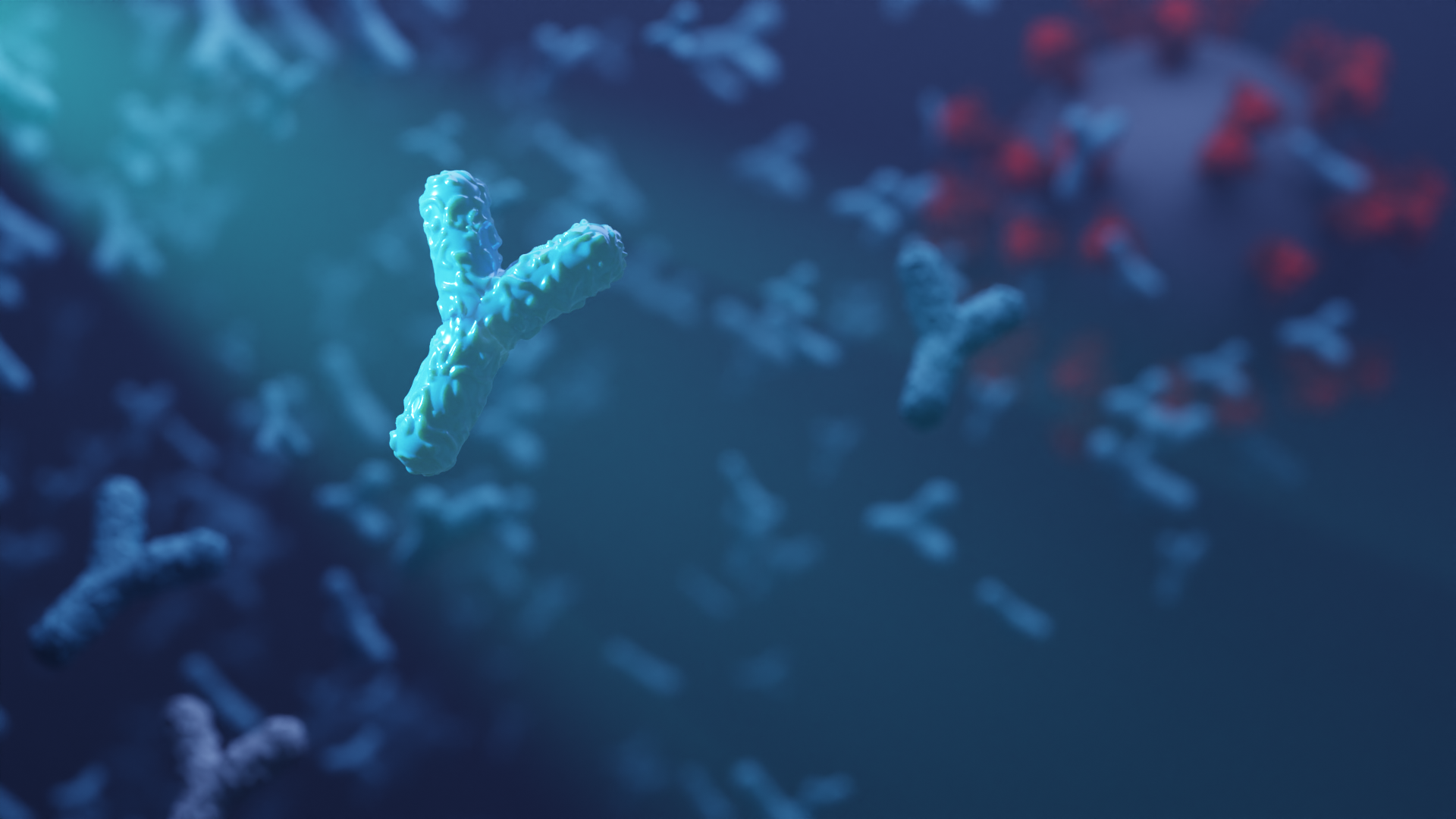
Custom biosensors for detecting coronavirus antibodies in blood
Today we report in Nature Biotechnology the design of custom protein-based biosensors that can detect coronavirus-neutralizing antibodies in blood. This research, which builds on prior sensor design technology developed in the Baker lab, was led by Baker lab postdoctoral scholars Jason Zhang, PhD, and Hsien-Wei (Andy) Yeh, PhD. From Behind the Paper: [W]e utilized the…
-

Rotory proteins designed from scratch
Today we report in Science the design of rotary devices made from custom proteins. These microscopic “axles” and “rotors” come together to form spinning assemblies, rather than being locked in just one orientation. Such mechanical coupling is a key feature of any machine. The new axle-rotor devices — which are each about a billion times…
-
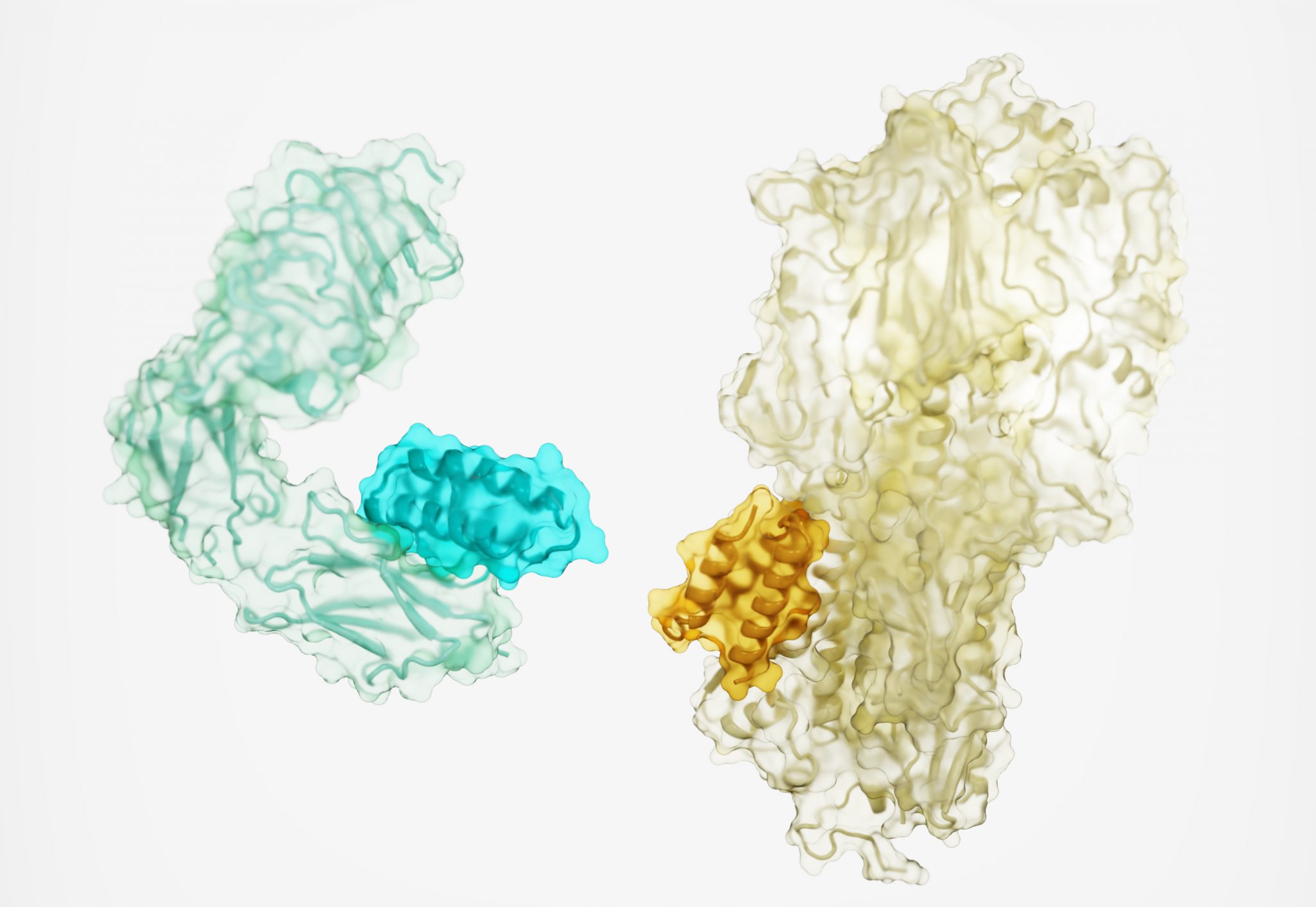
Protein drugs designed from the ground up
Today we report in Nature a new method for generating protein drugs. Using Rosetta-based design, an international team designed molecules that can target important proteins in the body, such as the insulin receptor, as well as proteins on the surface of viruses. This solves a long-standing challenge in drug development and may lead to new treatments for…
-
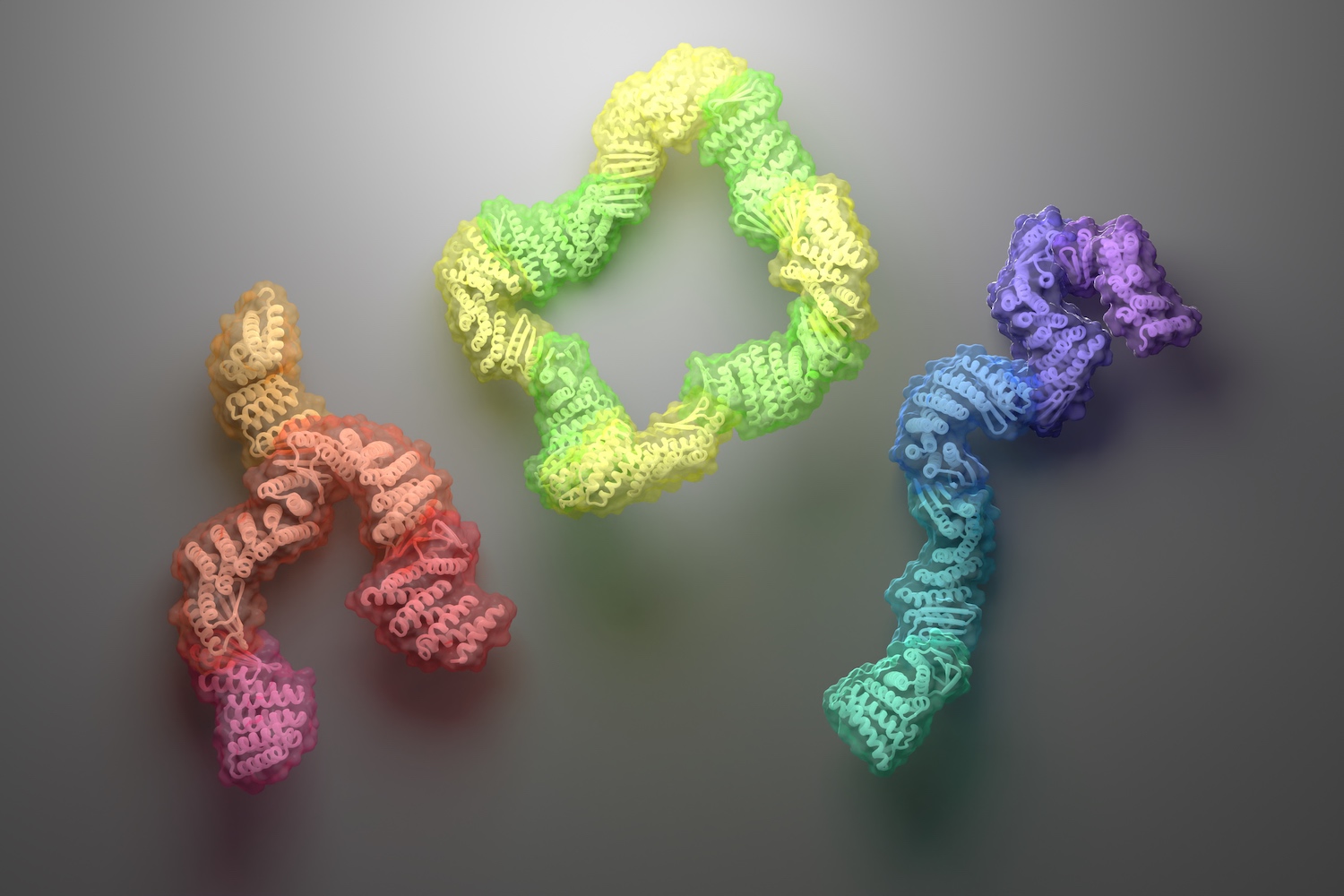
Diverse protein assemblies by (negative) design
A new approach for creating custom protein complexes yields asymmetric assemblies with interchangeable parts. Today we report in Science the design of new protein assemblies made from modular parts. These complexes — which adopt linear, branching, or closed-loop architectures — contain up to six unique proteins, each of which remains folded and soluble in the absence…
-

Deep learning dreams up new protein structures
Just as convincing images of cats can be created using artificial intelligence, new proteins can now be made using similar tools. In a new report in Nature, we describe the development of a neural network that “hallucinates” proteins with new, stable structures. “For this project, we made up completely random protein sequences and introduced mutations…
-
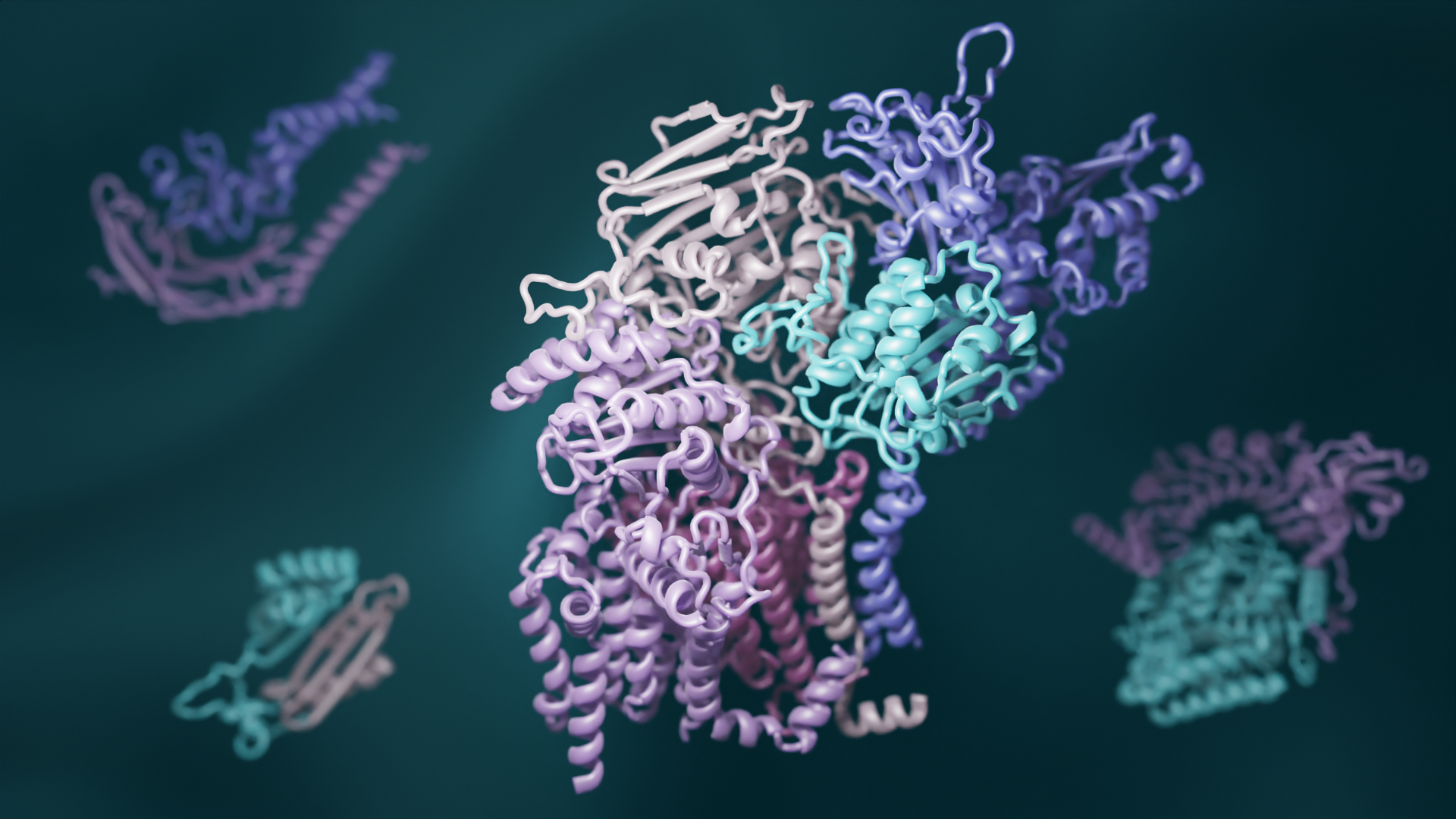
Deep learning reveals how proteins interact
A team led by scientsts in the Baker lab has combined recent advances in evolutionary analysis and deep learning to build three-dimensional models of how most proteins in eukaryotes interact. This breakthrough has significant implications for understanding the biochemical processes that are common to all animals, plants, and fungi. This open-access work appears in Science.…
-

RoseTTAFold: Accurate protein structure prediction accessible to all
Today we report the development and initial applications of RoseTTAFold, a software tool that uses deep learning to quickly and accurately predict protein structures based on limited information. Without the aid of such software, it can take years of laboratory work to determine the structure of just one protein. With RoseTTAFold, a protein structure can be…
-

Nanoparticle flu shot blocks seasonal and pandemic strains
IPD researchers together with collaborators at the National Institutes of Health have developed experimental flu shots that protect animals from a wide variety of seasonal and pandemic influenza strains. The lead vaccine candidate has entered human clinical testing at the NIH. If it proves safe and effective, these next-generation influenza vaccines may replace current seasonal…
-
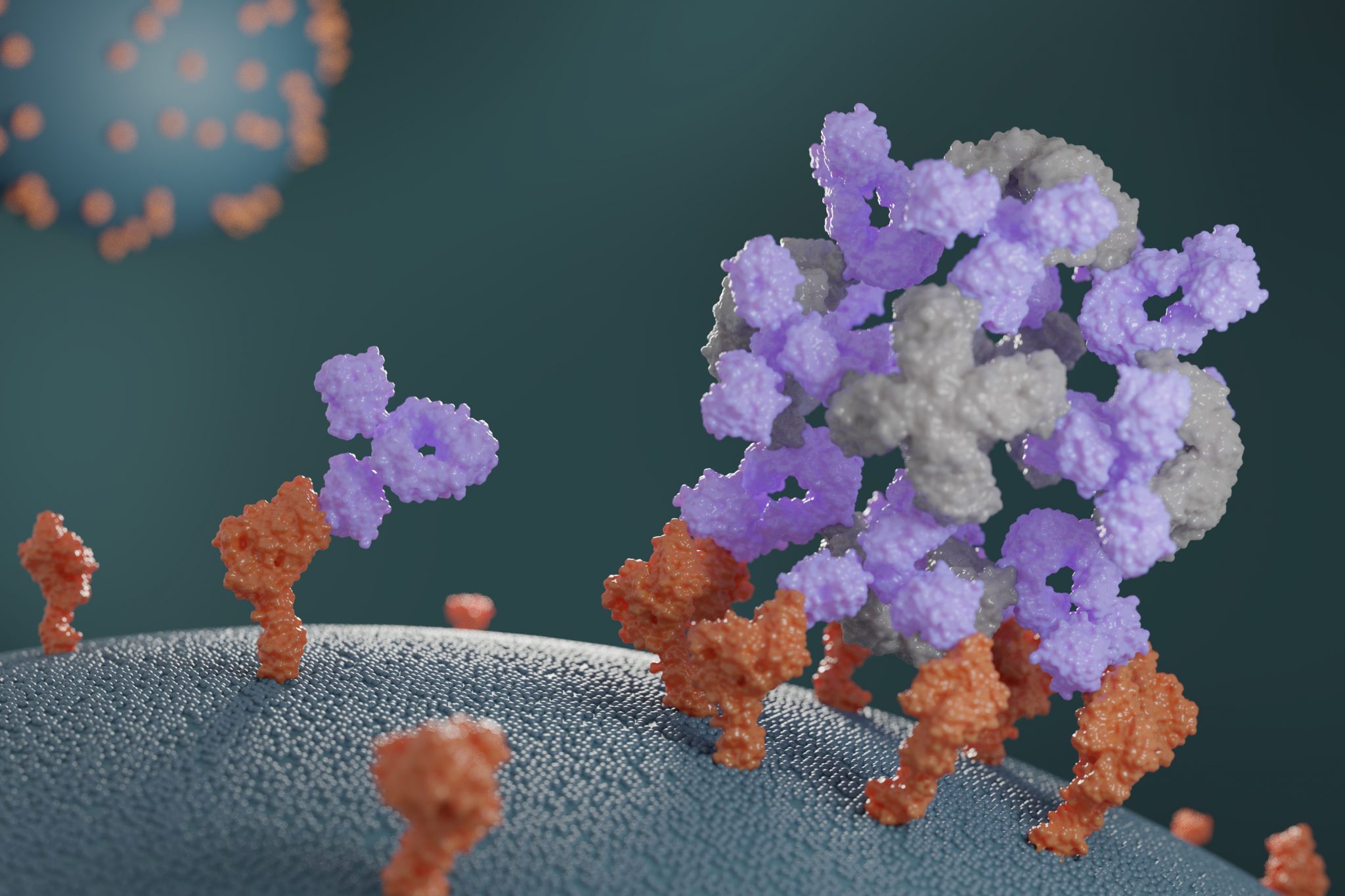
Companion proteins enhance antibody potency
This week we report the design of new proteins that cluster antibodies into dense particles, rendering them more effective. In laboratory testing, such clustered antibodies neutralize COVID-19 pseudovirus, enhance cell signaling, and promote the growth of T cells more effectively than do free antibodies. This new method for enhancing antibody potency may eventually be used…
-
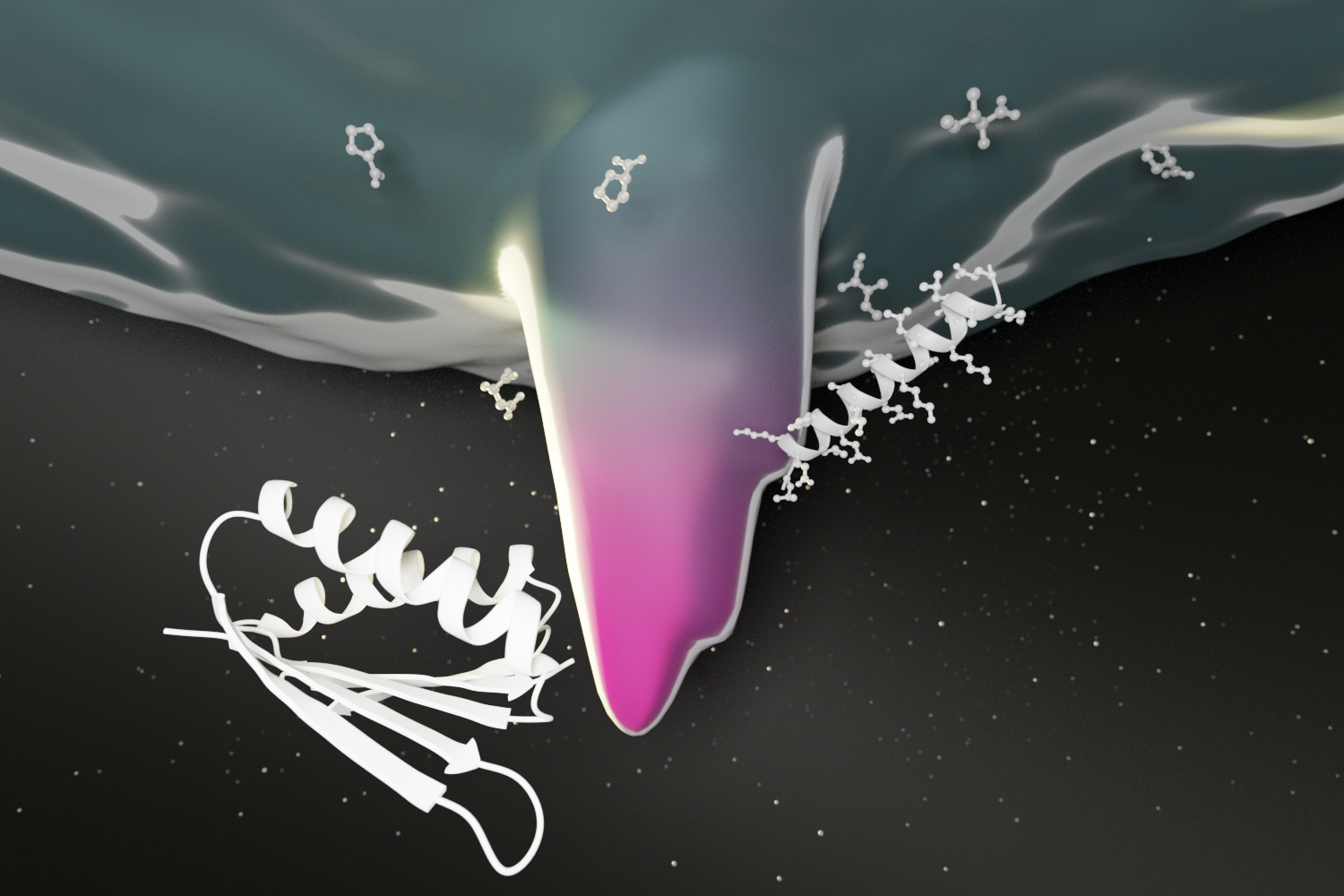
A deep learning approach to protein design
Scientists at the IPD and Ovchinnikov lab at Harvard have applied deep learning to the challenge of protein design, yielding a new way to quickly create protein sequences that fold up as desired. This breakthrough has broad implications for the development of protein-based medicines and vaccines. Computational protein design has primarily focused on finding amino…
-

Design of transmembrane beta barrels
Biochemists have created barrel-shaped proteins that embed into lipid membranes, expanding the bioengineering toolkit. In a milestone for biomolecular design, a team of scientists has succeeded in creating new proteins that adopt one of the most complex folds known to molecular biology. These designer proteins were shown in the lab to spontaneously fold into their…
-
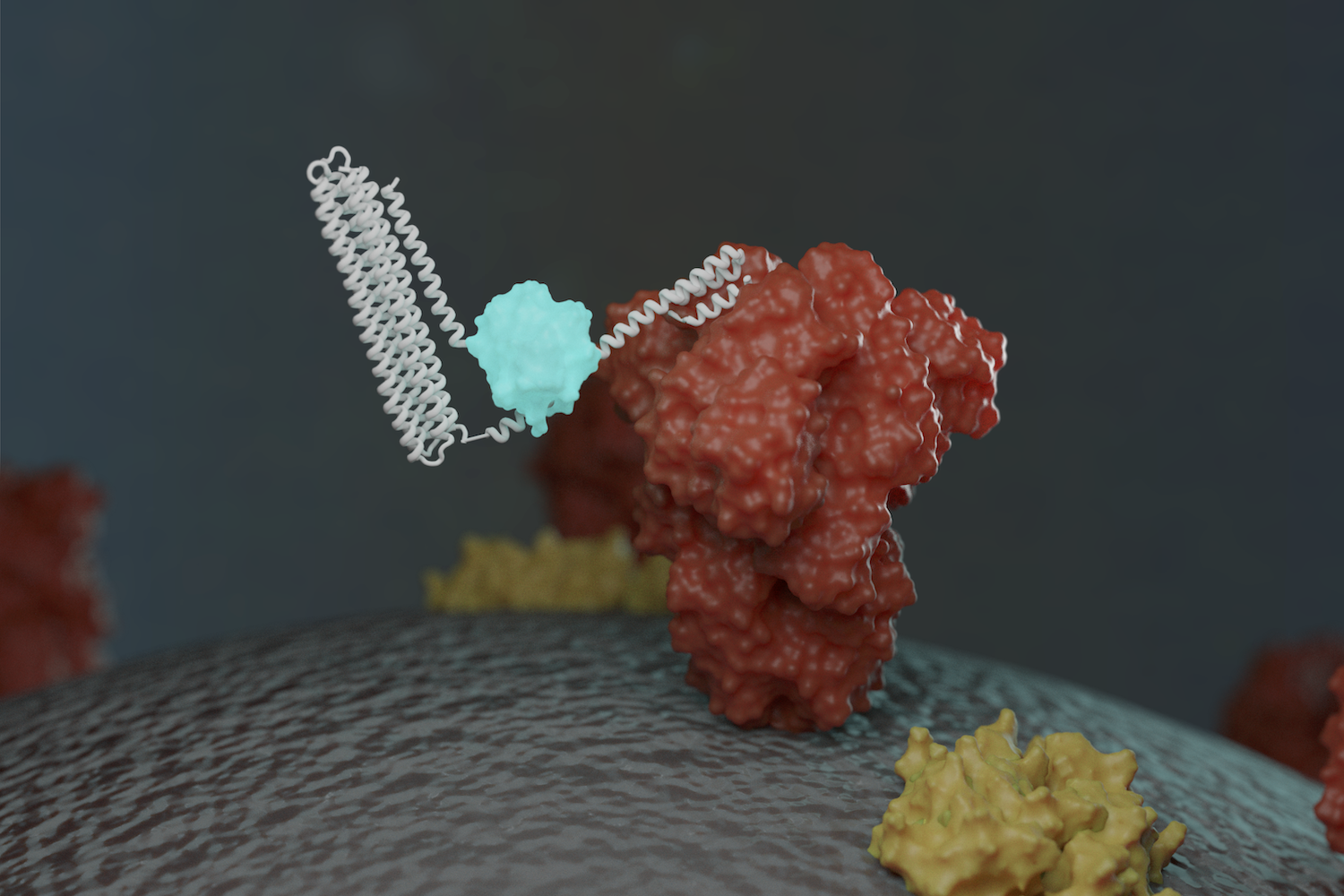
New sensors detect coronavirus proteins and antibodies
This week we report [PDF] a new way to detect the virus that causes COVID-19, as well as antibodies against it. Scientists at the Institute for Protein Design have created protein-based sensors that glow when mixed with components of the virus or specific antibodies. This breakthrough could enable faster and more widespread testing in the near…
-

Protein patches boost cell signaling
Today we report the design of a new class of protein material that interacts with living cells without being absorbed by them. These large, flat arrays built from multiple protein parts can influence cell signaling by clustering and anchoring cell surface receptors. This breakthrough could have far-reaching implications for stem cell research and enable the…
-
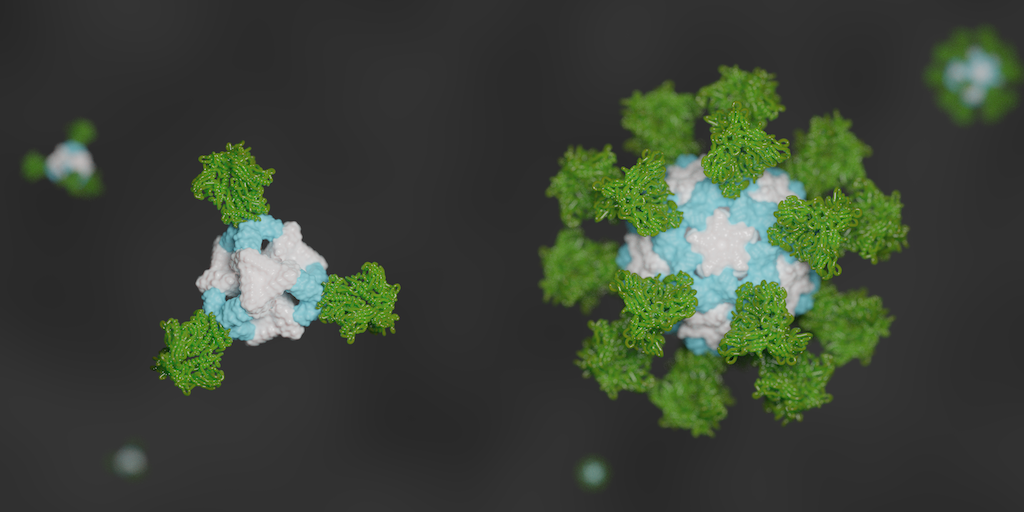
Design of an ultrapotent COVID-19 vaccine candidate
Today we report in Cell (PDF) the design and initial preclinical testing of an innovative nanoparticle vaccine candidate for the pandemic coronavirus. It produces virus-neutralizing antibodies in mice at levels ten-times greater than is seen in people who have recovered from COVID-19. Compared to vaccination with the soluble SARS-CoV-2 Spike protein, which is what many leading…
-
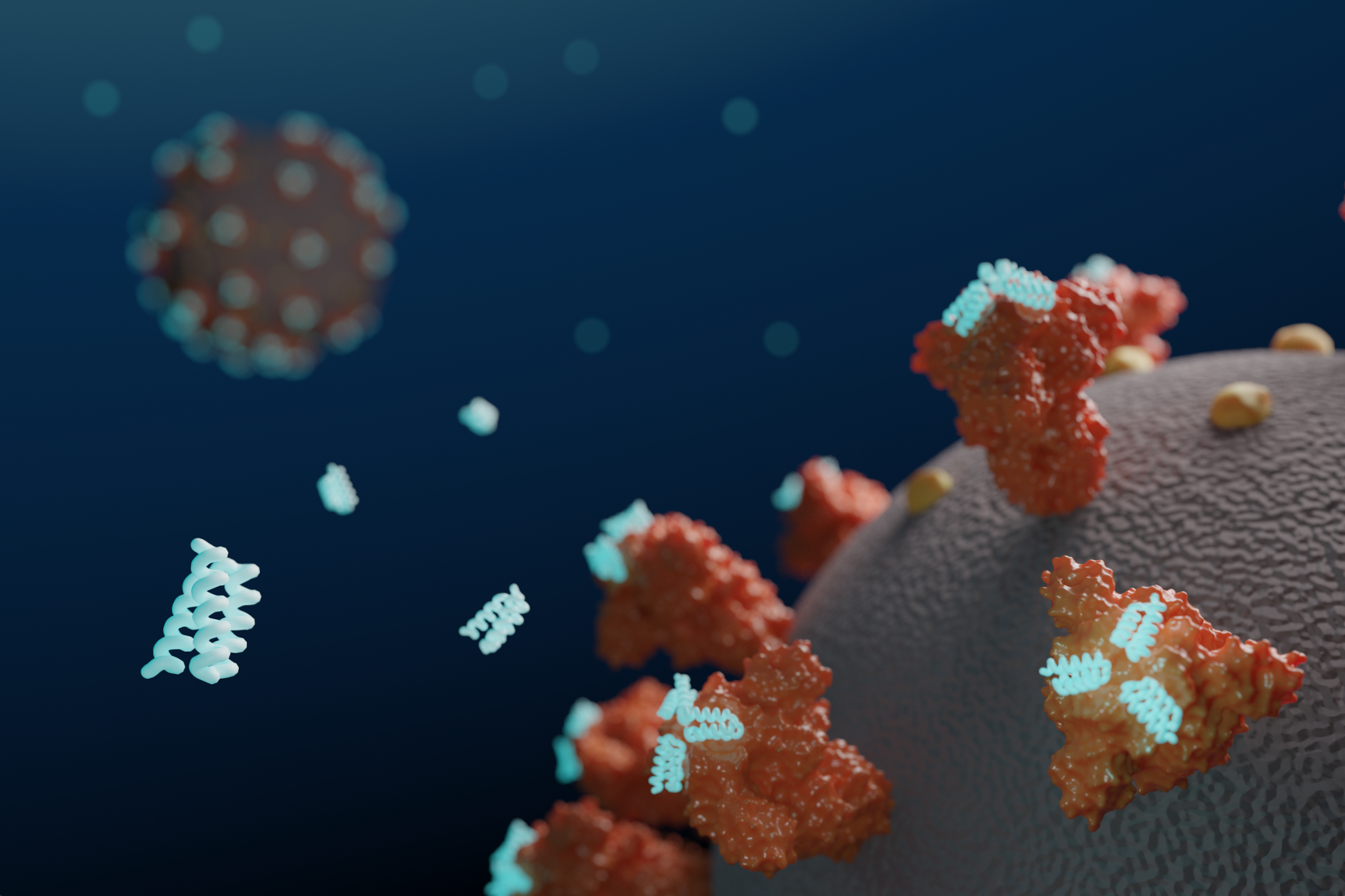
Antiviral proteins block coronavirus infection in the lab
Today we report in Science [PDF] the design of small proteins that protect cells from SARS-CoV-2, the virus that causes COVID-19. In experiments involving lab-grown human cells, the activity of the lead antiviral candidate produced (LCB1) was found to rival that of the best-known SARS-CoV-2 neutralizing antibodies. LCB1 is currently being evaluated in rodents. Coronaviruses…
-
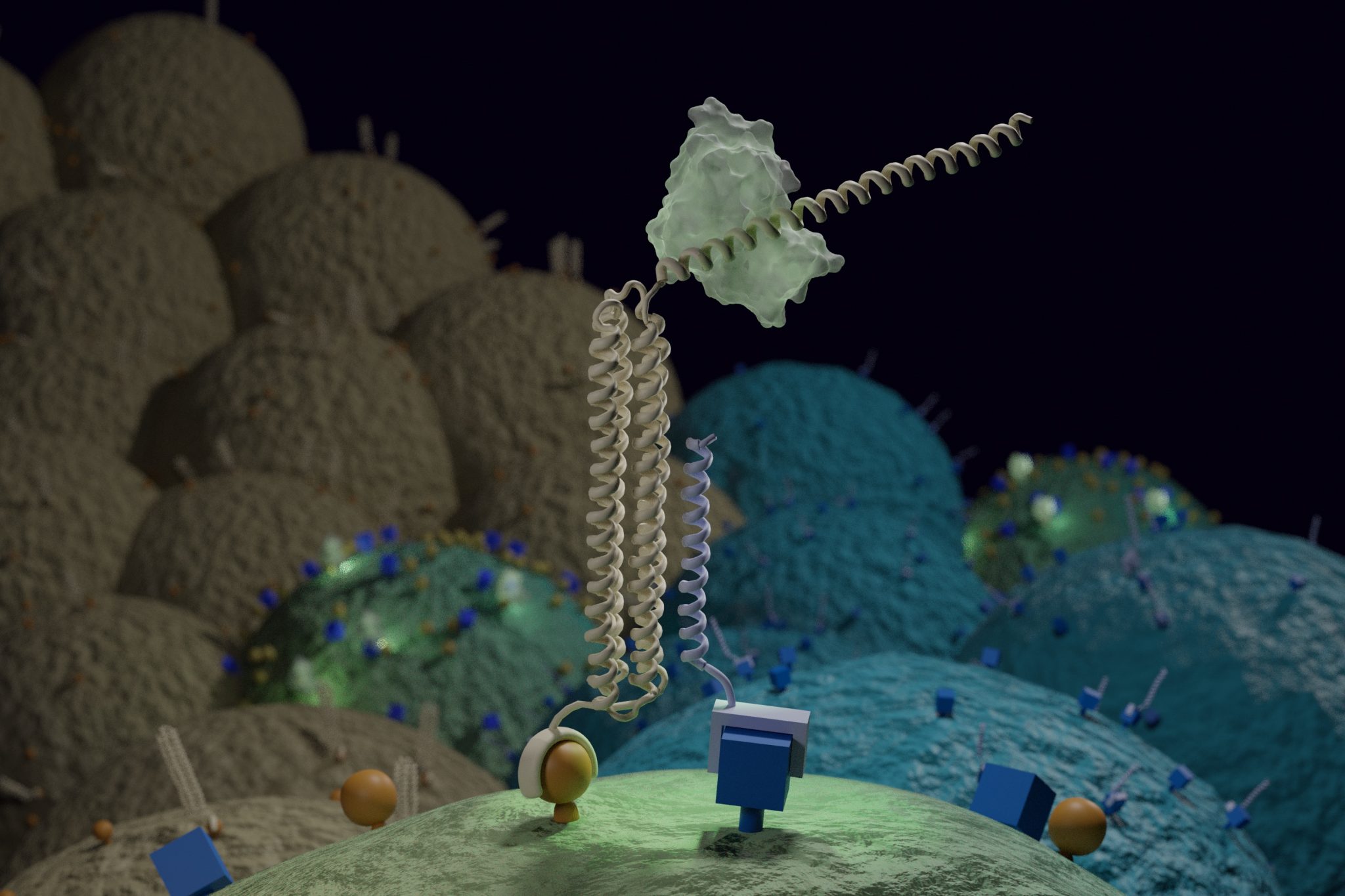
Introducing Co-LOCKR: designed protein logic for cell targeting
In a new paper [PDF] appearing in Science, a team of IPD researchers together with colleagues at UW Medicine and Fred Hutchinson Cancer Research Center demonstrate a new way to precisely target cells — including those that look almost exactly like their neighbors. They designed nanoscale devices made of synthetic proteins that target a therapeutic agent only to cells with…
-
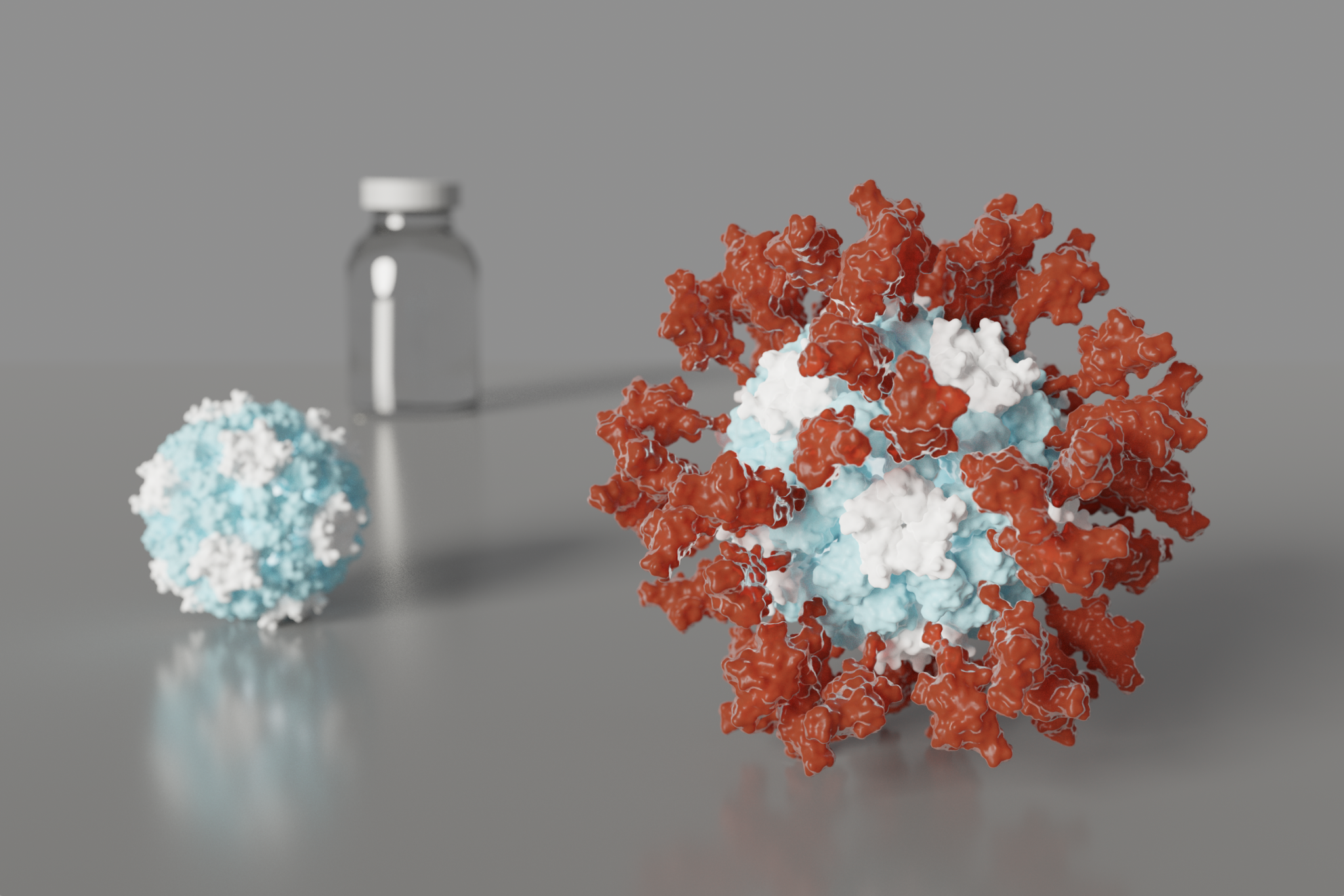
De novo nanoparticles as vaccine scaffolds
IPD researchers have developed a new vaccine design strategy that could confer improved immunity against certain viruses, including those that cause AIDS, the flu, and COVID-19. Using this technique, viral antigens are attached to the surface of self-assembling, de novo designed protein nanoparticles. This enables an unprecedented level of control over the molecular configuration of the resulting…
-
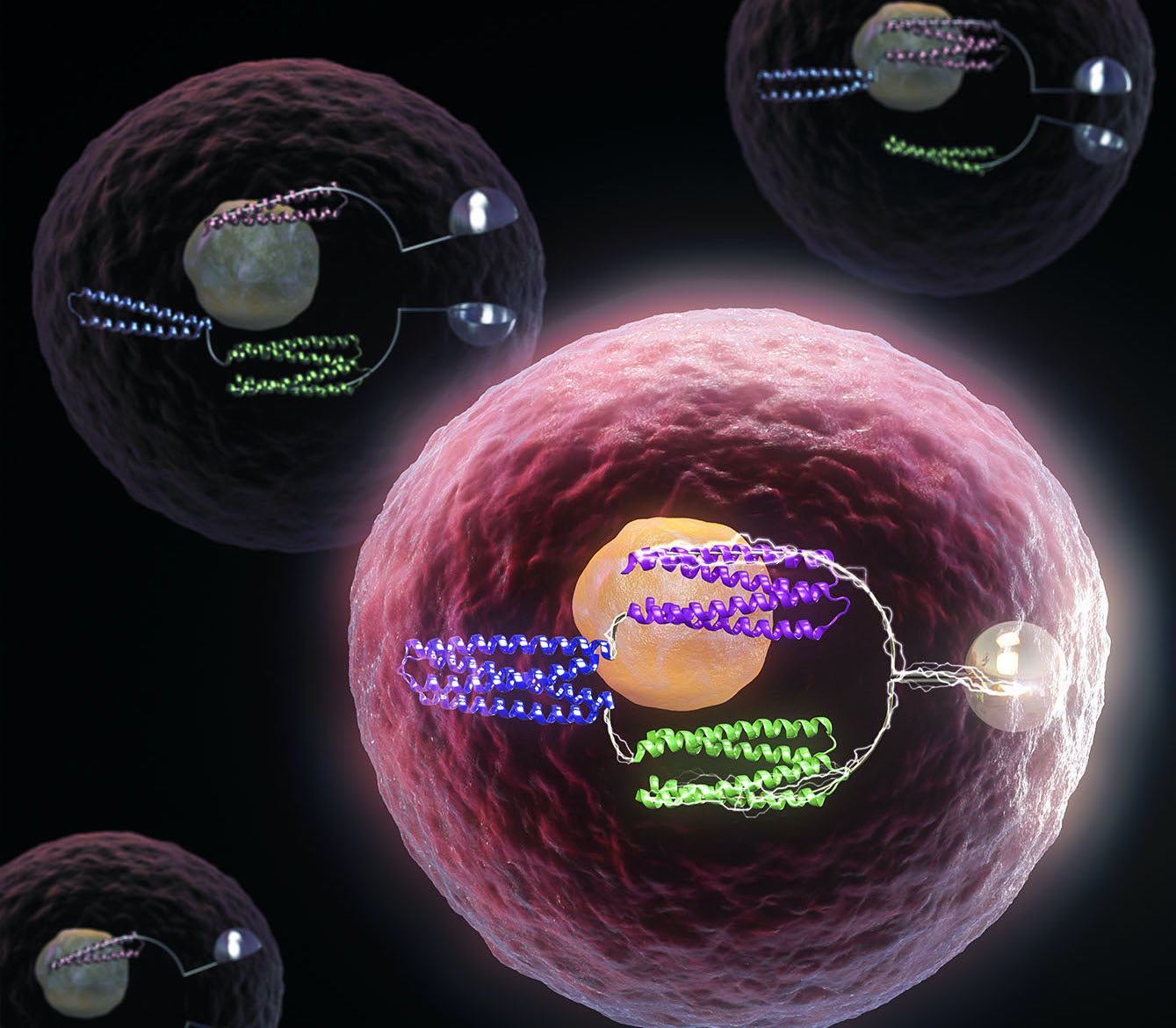
De novo design of protein logic gates
The same basic tools that allow computers to function are now being used to control life at the molecular level, with implications for future medicines and synthetic biology. Together with collaborators, we have created artificial proteins that function as molecular logic gates. These tools, like their electronic counterparts, can be used to program the behavior…
-
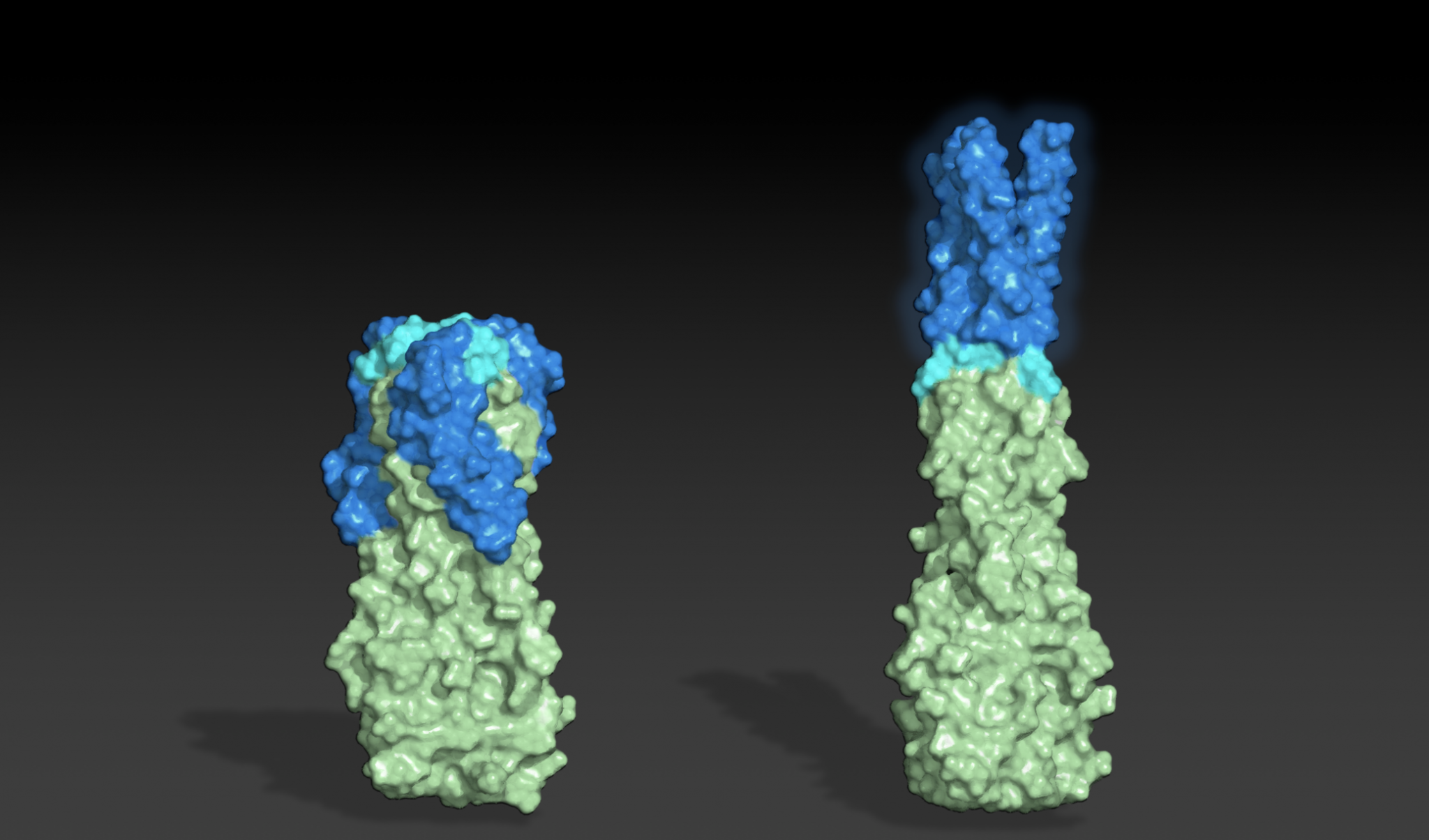
Designing shape-shifting proteins
Today we report the design of protein sequences that adopt more than one well-folded structure, reminiscent of viral fusion proteins. This research moves us closer to creating artificial protein systems with reliable moving parts. In nature, many proteins change shape in response to their environment. This plasticity is often linked to biological function. While computational…
-

Introducing LOCKR: a bioactive protein switch
Today we report in Nature the design and initial applications of the first completely artificial protein switch that can work inside living cells to modify—or even commandeer—the cell’s complex internal circuitry. The switch is dubbed LOCKR, short for Latching, Orthogonal Cage/Key pRotein. “In the same way that integrated circuits enabled the explosion of the computer chip industry, these versatile and dynamic…
-
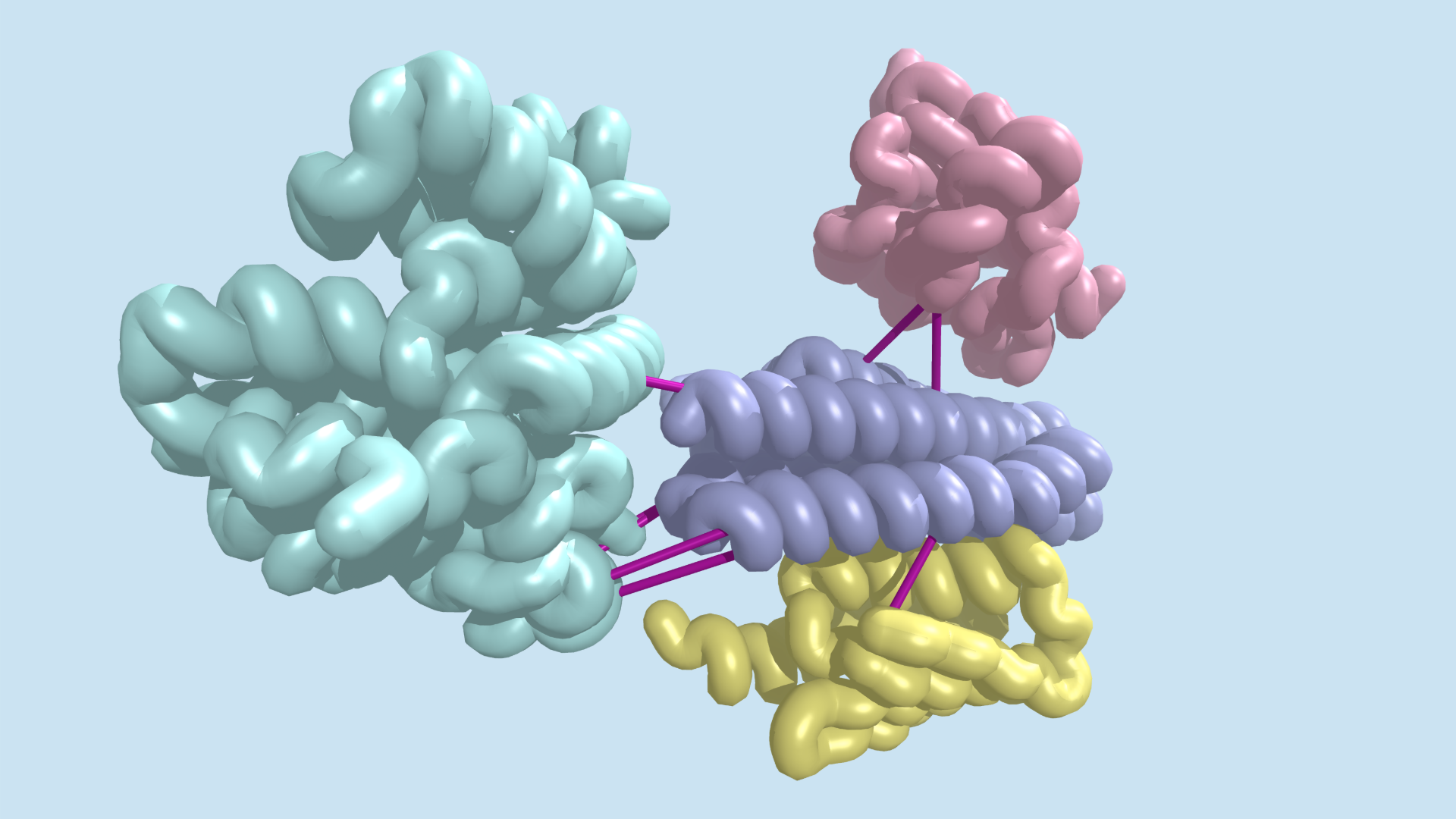
Coevolution at the proteome scale
Today we report in Science the identification of hundreds of previously uncharacterized protein–protein interactions in E. coli and the pathogenic bacterium M. tuberculosis. These include both previously unknown protein complexes and previously uncharacterized components of known complexes. This research was led by postdoctoral fellow Qian Cong and included former Baker lab graduate student Sergey Ovchinnikov, now a John Harvard Distinguished Science…
-

Protein arrays on mineral surfaces
Today we report the design of synthetic protein arrays that assemble on the surface of mica, a common and exceptionally smooth crystalline mineral. This work, which was performed in collaboration with the De Yoreo lab at PNNL, provides a foundation for understanding how protein-crystal interactions can be systematically programmed. Our goal was to engineer artificial proteins to self-assemble on…
-
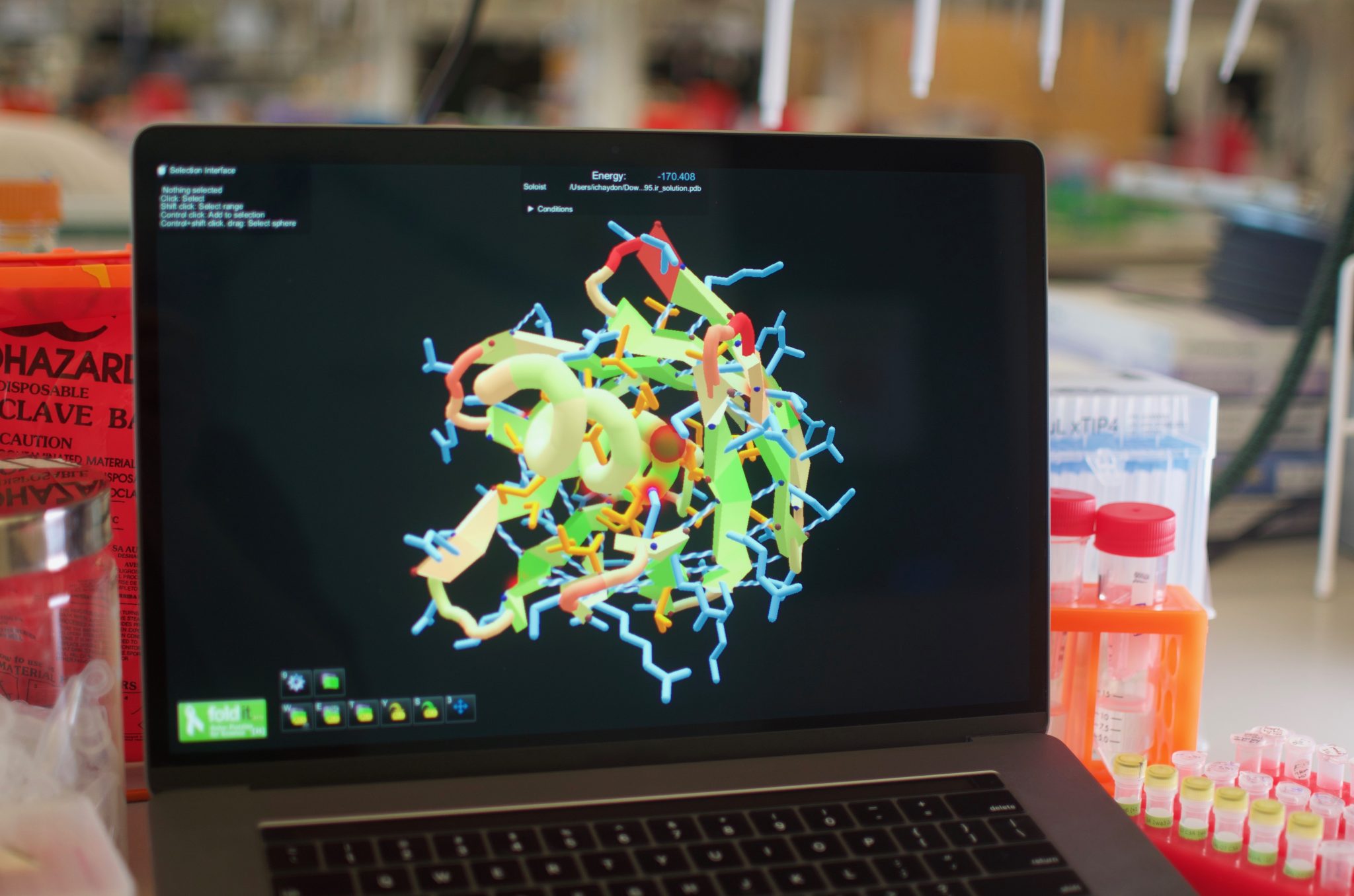
Protein design by citizen scientists
Citizen scientists can now use Foldit to successfully design synthetic proteins. The initial results of this unique collaboration appear today in Nature. Brian Koepnick, a recent PhD graduate in the Baker lab, led a team that worked on Foldit behind the scenes, introducing new features into the game that they believed would help players home in on…
-
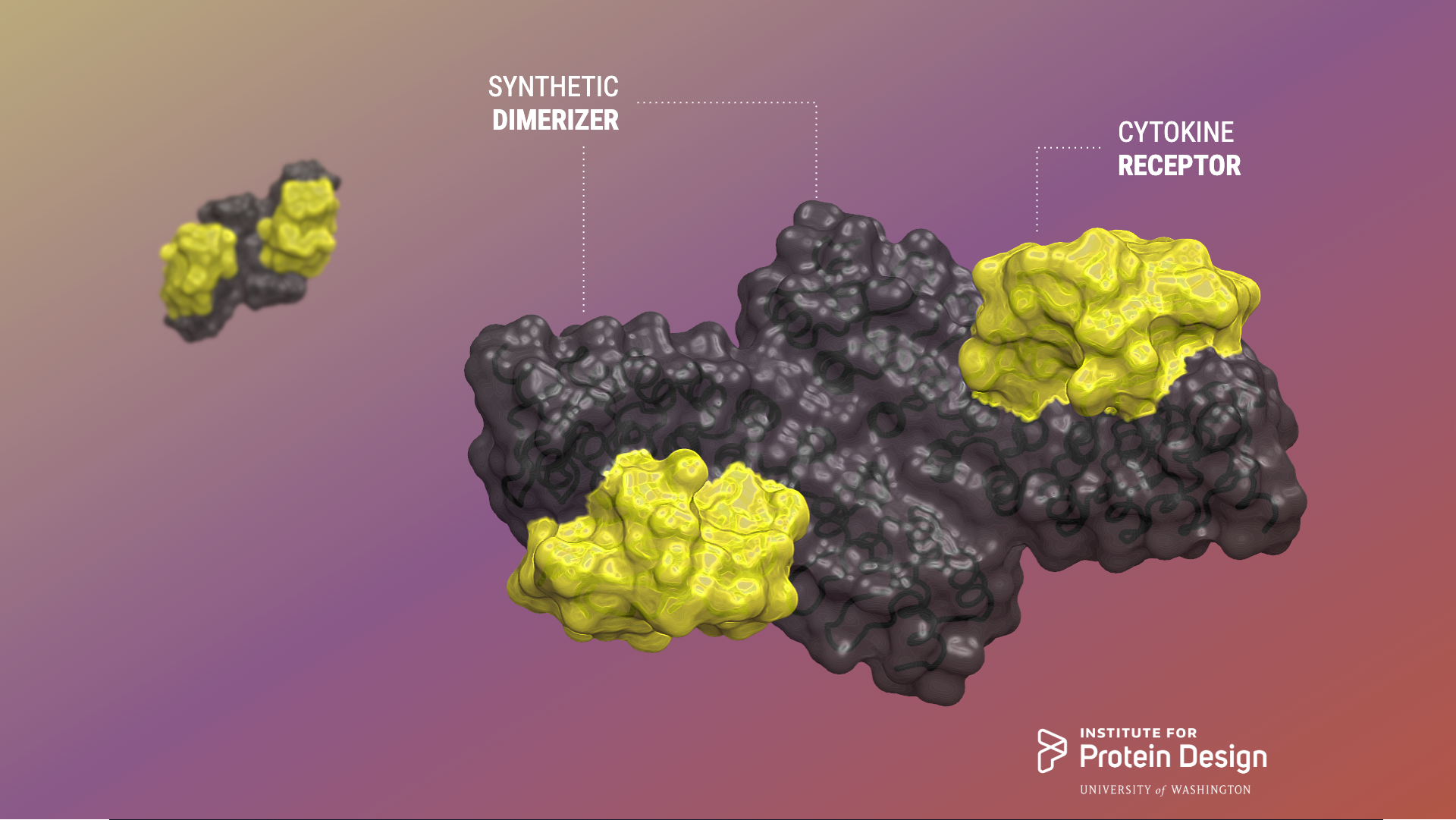
Designed ligands tune cytokine signaling
Today the Baker lab shares some exciting collaborative results of their efforts to design rigid and tunable receptor dimerizers. The first authors of this report are Kritika Mohan, Stanford, and George Ueda, IPD. From Science: Exploring a range of signaling Cytokines are small proteins that bind to the extracellular domains of transmembrane receptors to activate signaling pathways…
-
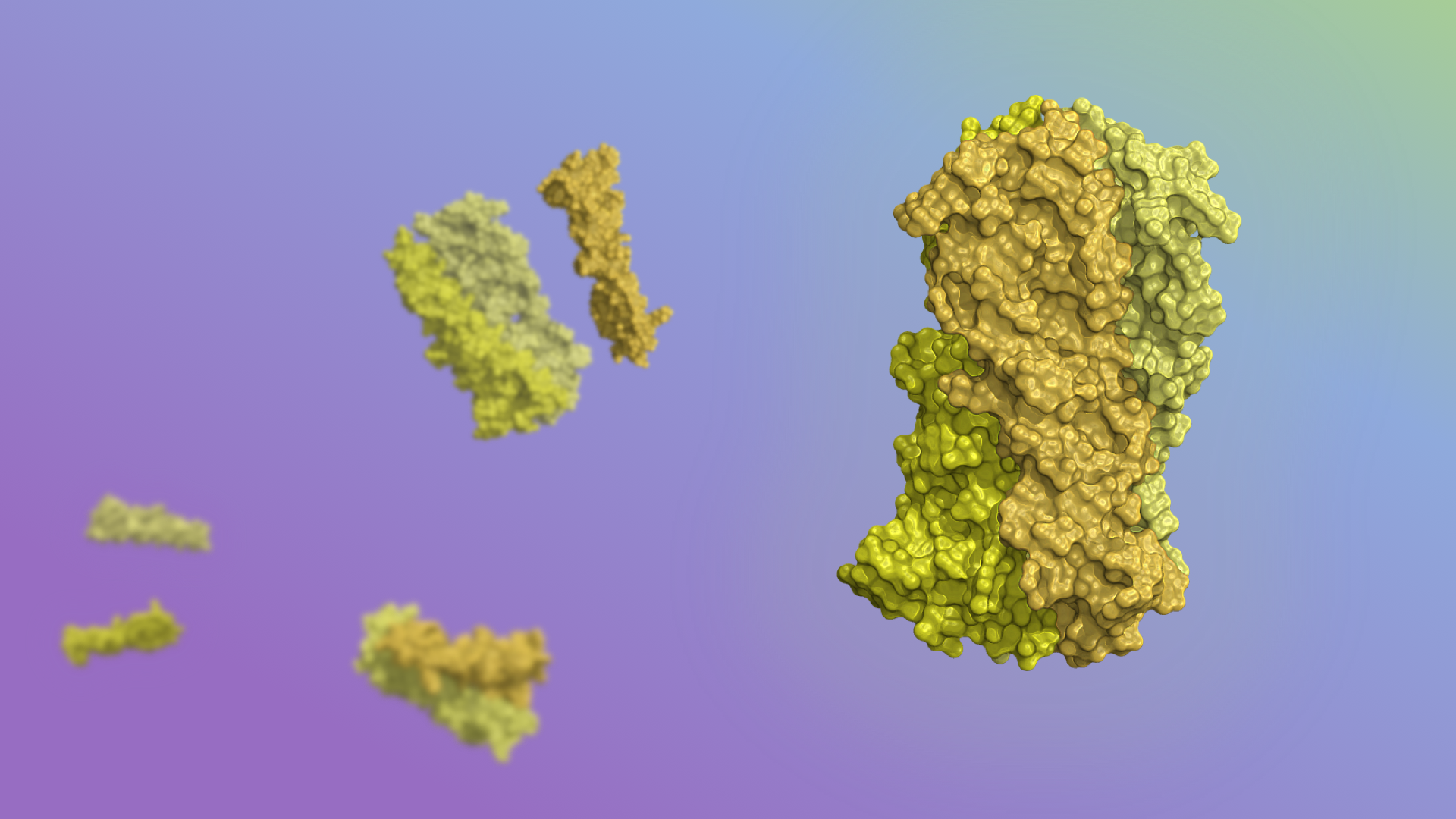
Tunable pH-dependent assemblies
Natural proteins often shift their shapes in precise ways in order to function. Achieving similar molecular rearrangements by design, however, has been a long-standing challenge. Today, a team of researchers lead by scientists at the IPD report in Science the rational design of synthetic proteins that move in response to their environment in predictable and tunable…
-
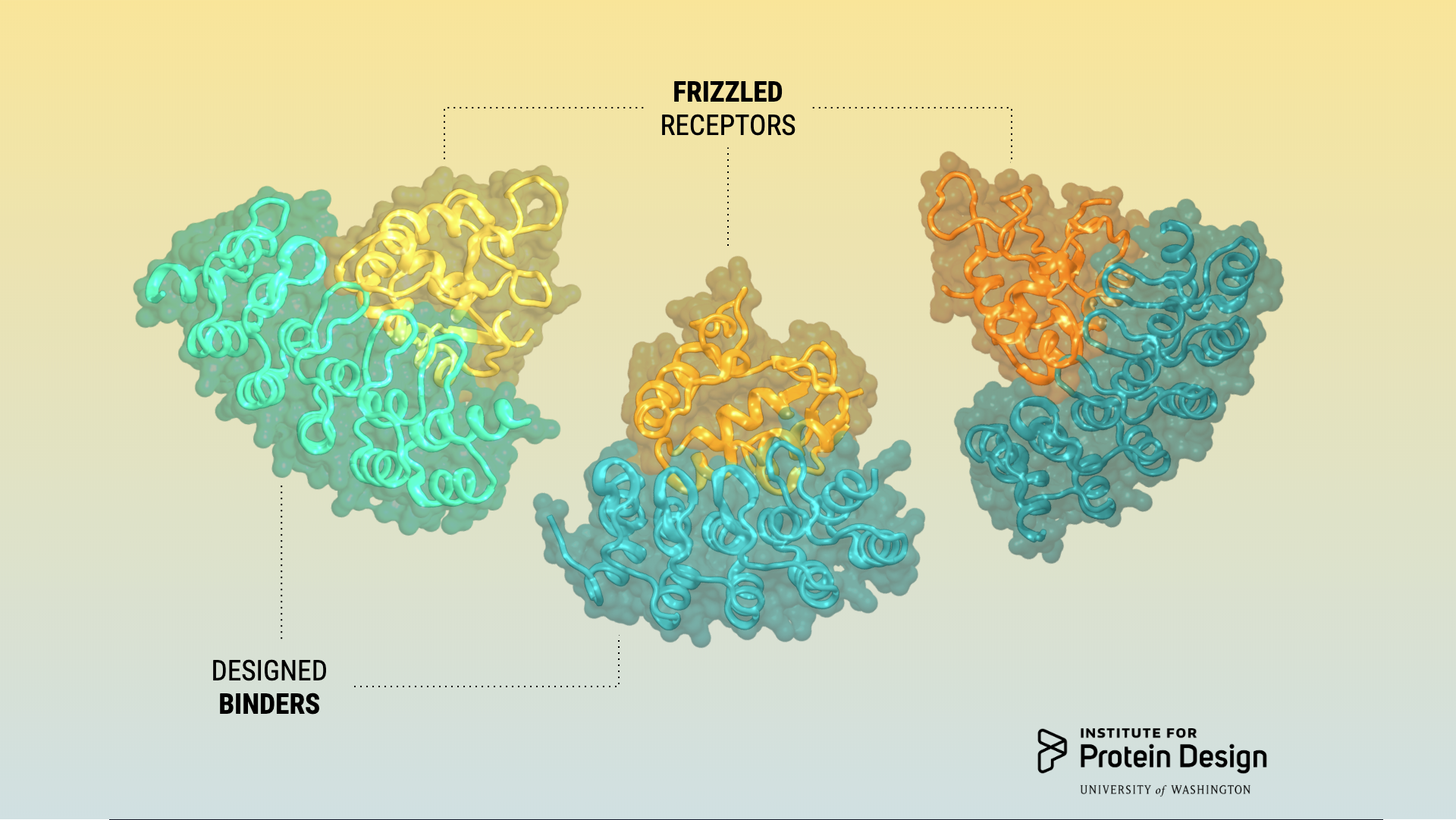
Receptor sub-type binders
This week we report in NSMB a combined computational design and experimental selection approach for creating proteins that bind selectively to closely related receptor subtypes. This project was led by Luke Dang, a former Baker lab graduate student, and Yi Miao, a postdoctoral researcher in Christopher Garcia’s lab at Stanford. Abstract: To discriminate between closely related members of…
-
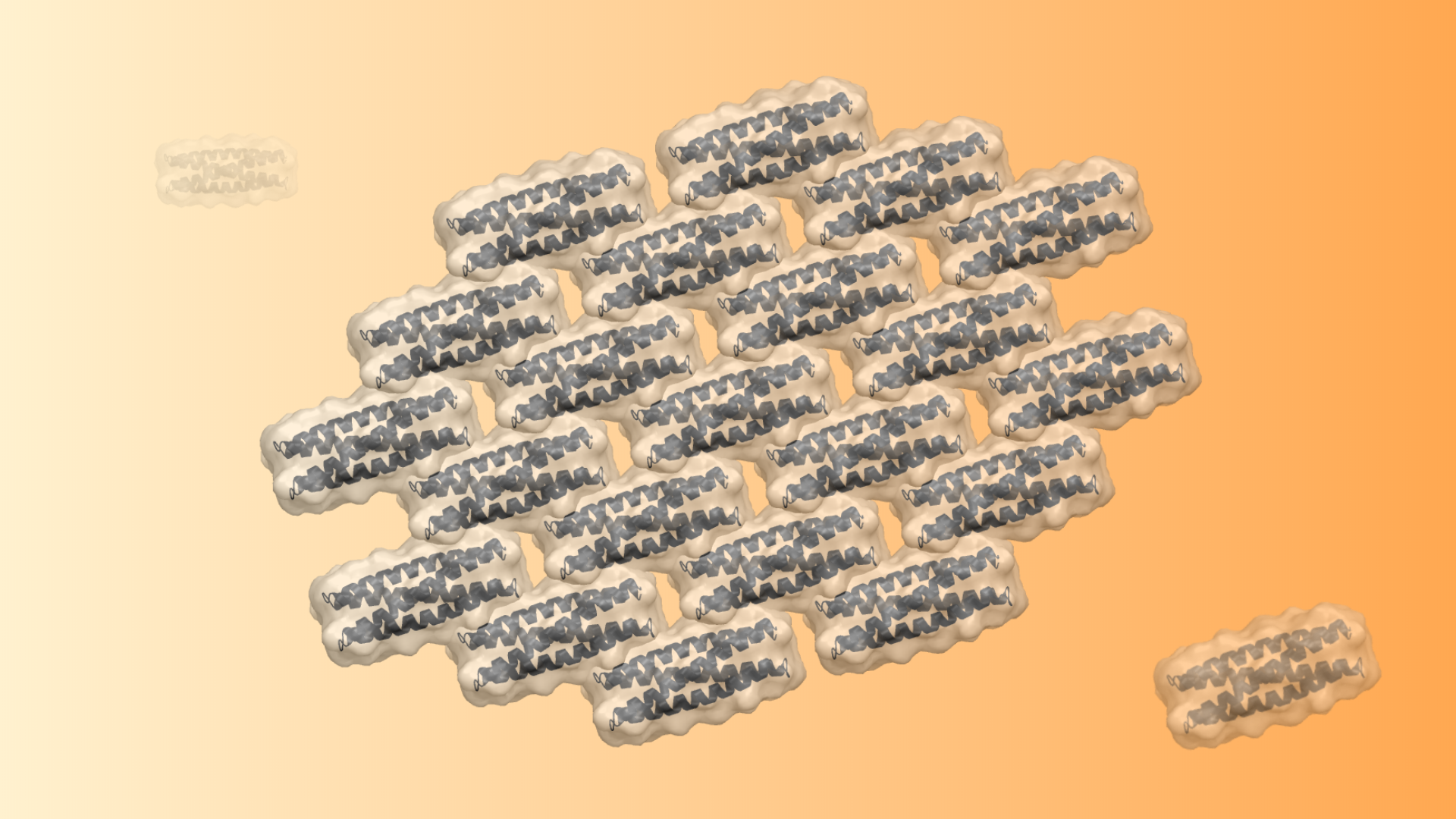
De novo 2D arrays
This week we report in JACS a general approach for designing self-assembling 2D protein arrays. This project was led by Zibo Chen, a recent Baker lab graduate student, and featured collaborators from the, DiMaio, De Yoreo and Kollman labs at UW. Abstract: Modular self-assembly of biomolecules in two dimensions (2D) is straightforward with DNA but…
-
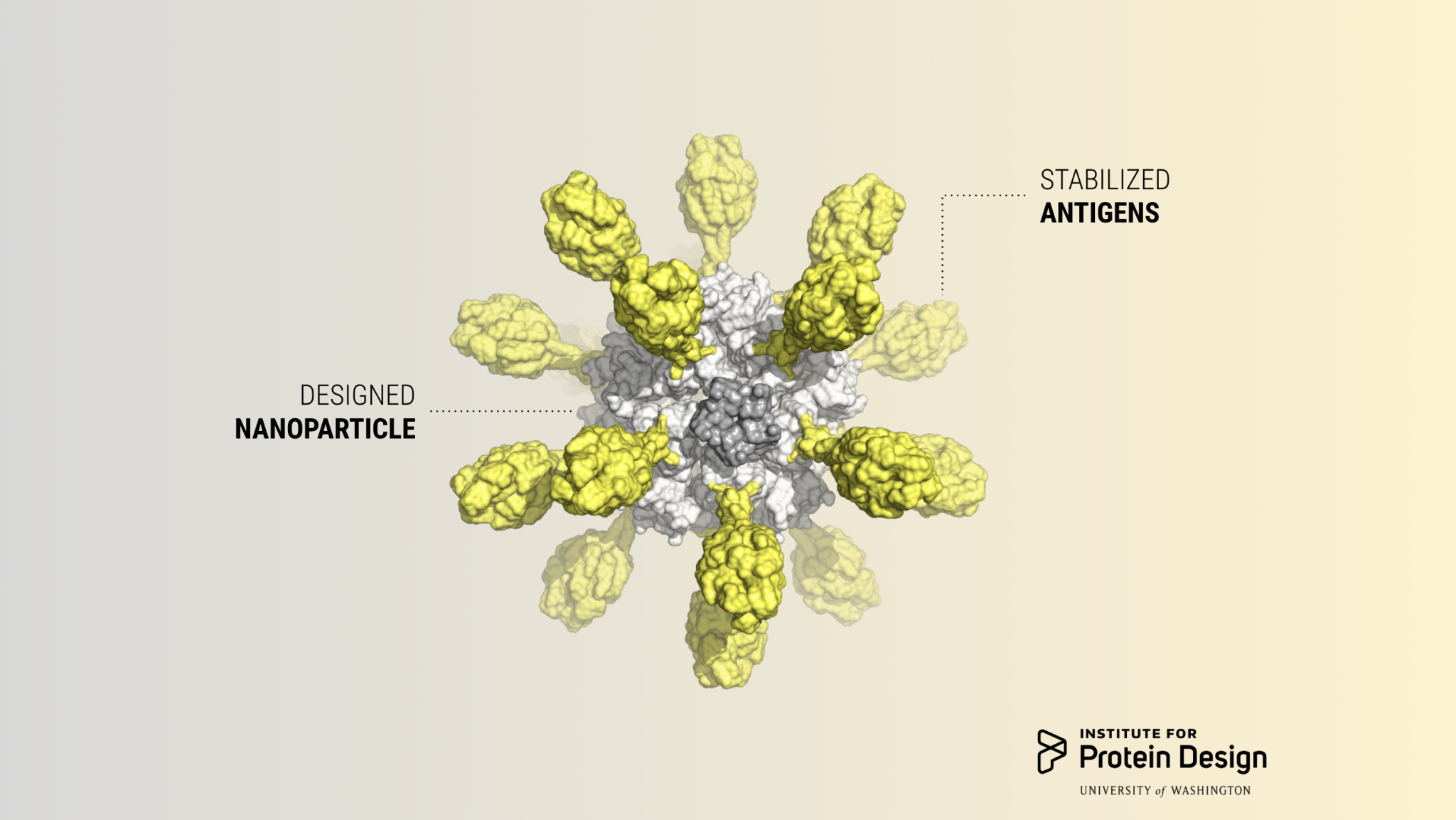
Designing a stable and potent RSV vaccine candidate
Today we report in Cell our first computer-designed nanoparticle vaccine targeting respiratory syncytial virus, the primary cause of pneumonia in young children and the leading cause of infant mortality worldwide after malaria. Although virtually every child will get infected by RSV before the age of three, an estimated 99 percent of deaths associated with the…
-
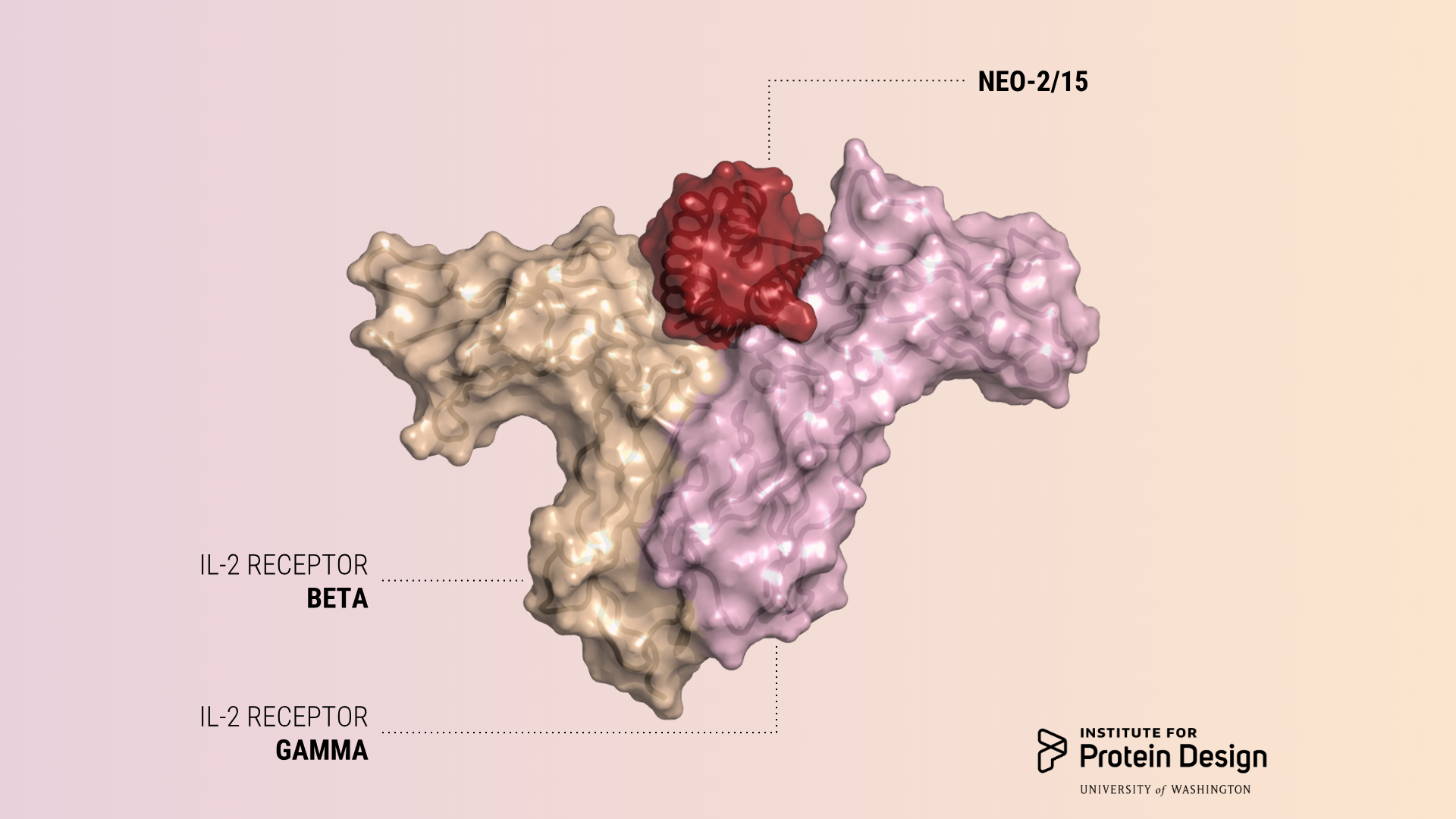
Potent anti-cancer proteins with fewer side effects
Today we report in Nature the first de novo designed proteins with anti-cancer activity. These compact molecules were designed to stimulate the same receptors as IL-2, a powerful immunotherapeutic drug, while avoiding unwanted off-target receptor interactions. We believe this is just the first of many computer-generated cancer drugs with enhanced specificity and potency. “People have tried for 30…
-
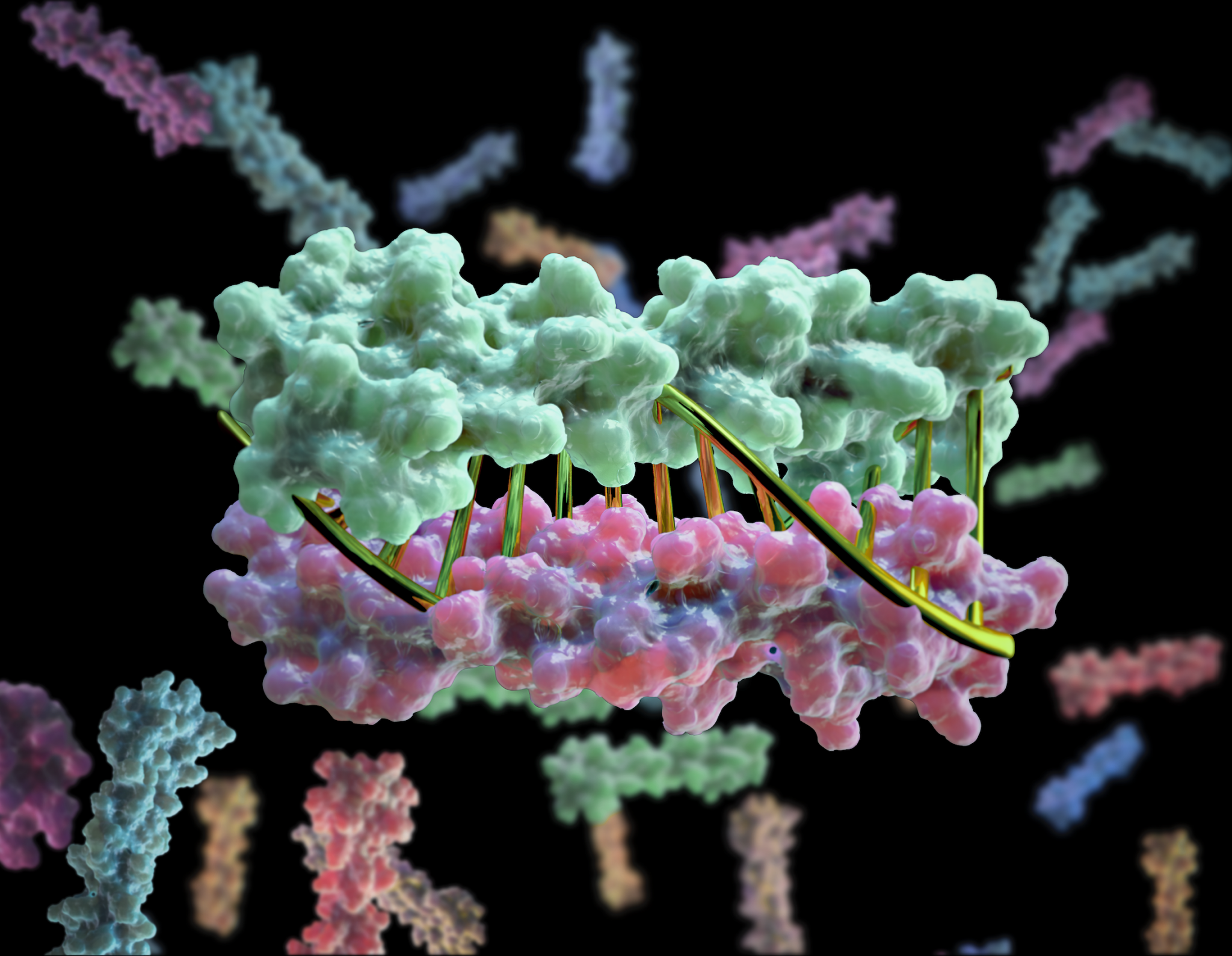
New designer proteins mimic DNA
To close out the year, Baker Lab scientists published a new report describing the creation of proteins that mimic DNA. We believe this breakthrough will aid the creation of bioactive nanomachines. DNA is a widely used building material at the nanoscale because it is simple and predictable: A pairs with T and C pairs with…
-
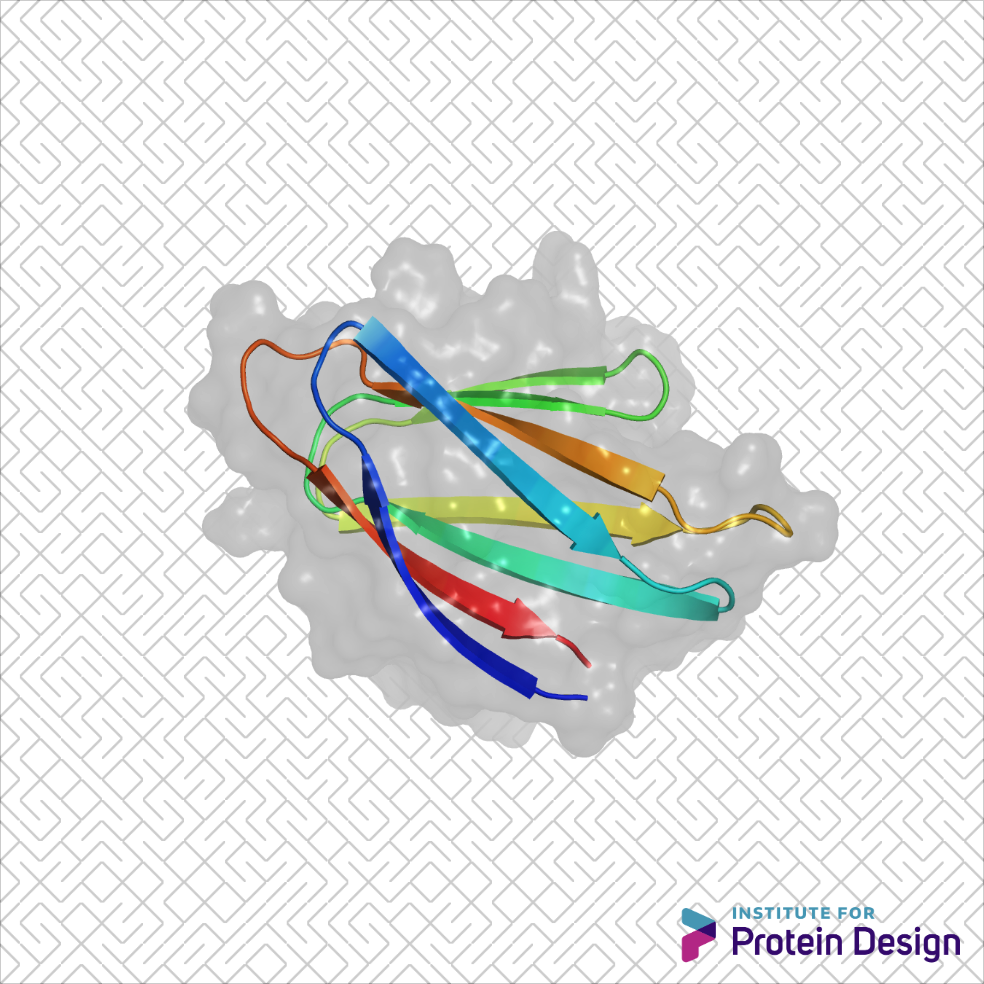
Rolling out new jellies
The basic parts of proteins — helices, strands and loops — can be combined in countless ways. But certain combinations are trickier than others. This week scientists from the IPD, along with collaborators in Brno and Santa Cruz, published the first-ever example of designed non-local beta-strand interactions. Beta-sheet proteins carry out critical functions in biology,…
-

Fluorescent proteins designed from scratch
In the summer of 1961, Osamu Shimomura drove across the country in a cramped station wagon to scoop jellyfish from the docks of Friday Harbor. He wanted to discover what made them glow. It took Shimomura and other biochemists more 30 years to find a full answer. By then, recombinant DNA technology allowed researchers to…
-
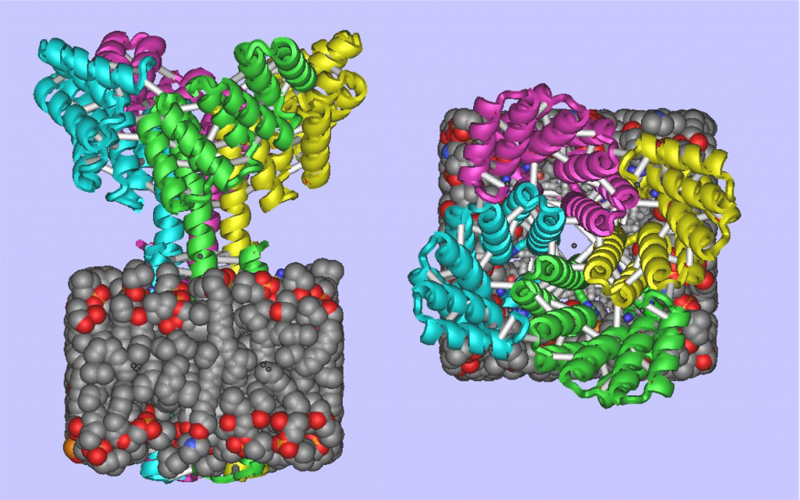
De Novo Design of Membrane Proteins
It is now possible to create complex, custom-designed transmembrane proteins from scratch ! Today Baker lab members published in Science “Accurate computational design of multipass transmembrane proteins” The Abstract reads as follows: The computational design of transmembrane proteins with more than one membrane-spanning region remains a major challenge. We report the design of transmembrane monomers,…
-
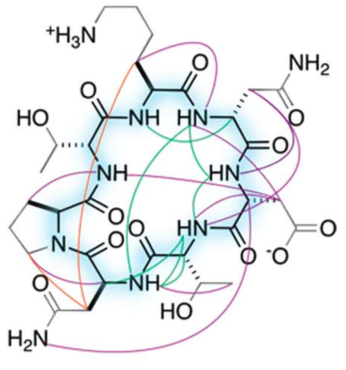
A New World of Designed Macrocycles
Today marks another major step forward for peptide based drug discovery. IPD researchers report in Science the computational design of a new world of small cyclic peptides, “Macrocycles”, increasing the number of the known kinds of these molecules by multiple fold. The conceptual art image below “Illuminating the energy landscape” shows the power of computational design…
-
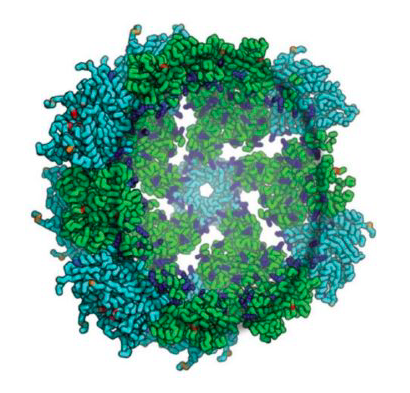
Synthetic Nucleocapsids Have Arrived
Published today in Nature, IPD researchers describe the first synthetic protein assemblies — dubbed synthetic nucleocapsids — that encapsulate their own genome and evolve in complex environments. Synthetic nucleocapsids are built to resemble viral capsids and could be used in future to deliver therapeutics to specific cells and tissues. These icosahedral protein assemblies are based off of…
-

Designs on New World of Mini-Protein Therapeutics
Mark your calendars! September 27, 2017 is the day the doors opened to whole new world of targeted therapeutics. The Baker lab and numerous talented collaborators published in Nature that it is now possible to conduct “Massively parallel de novo protein design for targeted therapeutics”. Three factors make this possible: Rosetta molecular modeling algorithms for computational protein…
-
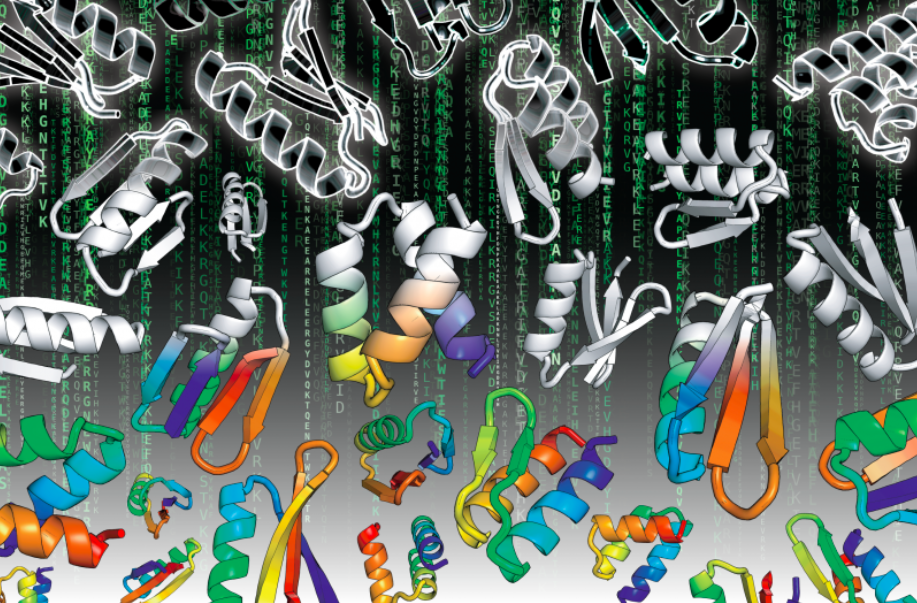
The Matrix of Protein Design
The Matrix movie (1999) depicts a future in which the reality perceived by most humans is actually a computer simulated reality called “the Matrix”. Published today in Science, the Baker lab and collaborators report on a new kind of Matrix – a new reality for large scale computational protein design which can achieve massive data driven improvements in our ability to design highly stable,…
-
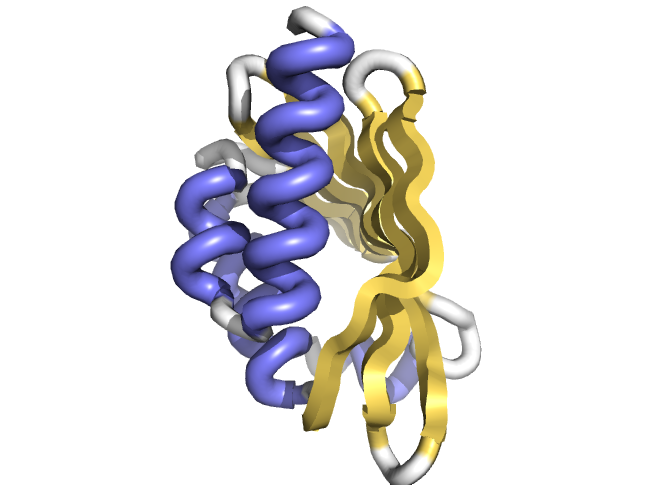
Design of novel cavity containing proteins
The latest paper coming out from the IPD was published today on the Science website. It’s titled “Principles for designing proteins with cavities formed by curved β sheets” with first co-authors Enrique Marcos and Benjamin Basanta, a former and current IPD member, respectively. Other IPD members on the paper include Tamuka Chidyausiku, Gustav Oberdorfer, Daniel-Adriano…
-

Unleashing the Power of Synthetic Proteins
Published today in Science Philanthropy Alliance, David Baker, Director of the Institute for Protein Design describes how the opportunities for computational protein design are endless — with new research frontiers and a huge variety of practical applications to be explored, from medicine to energy to technology. This is an exciting time as we are undergoing a technological revolution…
-

Hyper-stable Designed Peptides and the Coming of Age for De Novo Protein Design
Small constrained peptides combine the stability of small molecule drugs with the selectivity and potency of antibody-based therapeutics. However, peptide-based therapeutics have largely remained underexplored due to the limited diversity of naturally occurring peptide scaffolds, and a lack of methods to design them rationally. New computational design and wet lab methods developed at the Institute for…
-
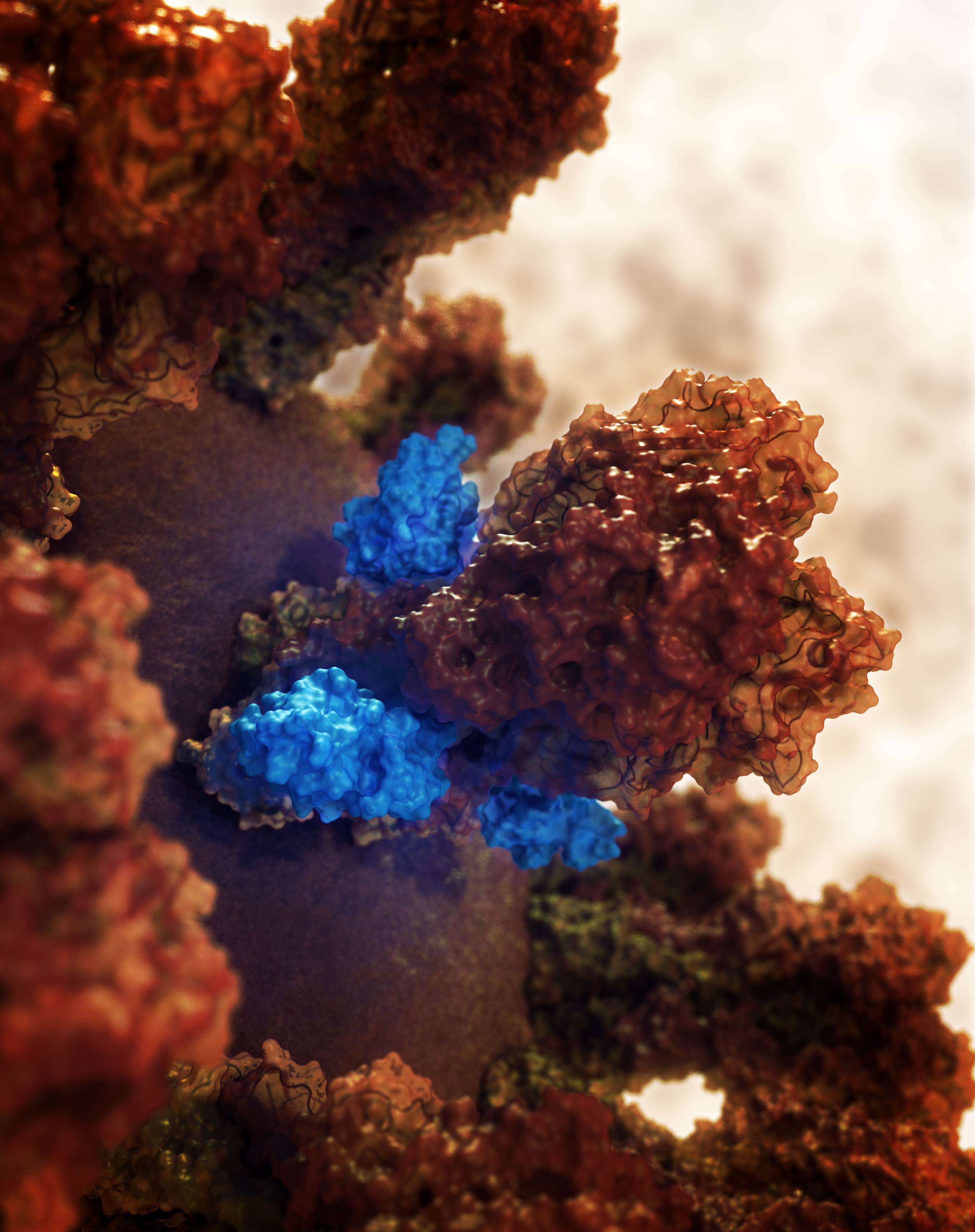
Designed Protein Containers Push Bioengineering Boundaries
Earlier this month, Baker lab researchers reported the computational design of a hyperstable 60-subunit protein icosahedron in Nature (Hsia et al); icosahedral protein structures are commonly observed in natural biological systems for packaging and transport (e.g. viral capsids). The described design was composed of 60 trimeric protein building blocks that self-assembled in a nanocage. In…
-
Flu Binder Paper Debuts in PLOS Pathogens
Today at 11am, the paper titled “A Computationally Designed Hemagglutinin Stem-Binding Protein Provides In Vivo Protection from Influenza Independent of a Host Immune Response” was published to the PLOS Pathogen website. This paper was contributed to by several IPD members, including Aaron Chevalier, Jorgen Nelson, Lance Stewart, Lauren Carter, and David Baker. The research was performed…
-
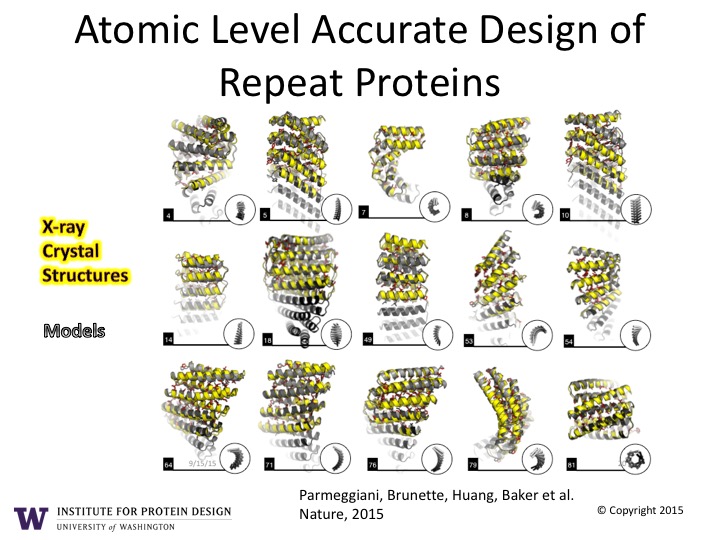
Big moves in protein structure prediction and design
[envira-gallery id=”3246″] Custom design with atomic level accuracy enables researchers to craft a whole new world of proteins Naturally occurring proteins are the nanoscale machines that carry out essentially all of the critical functions in living things. While it has been known for over 40 years that the sequence of amino acids completely determines the…
-
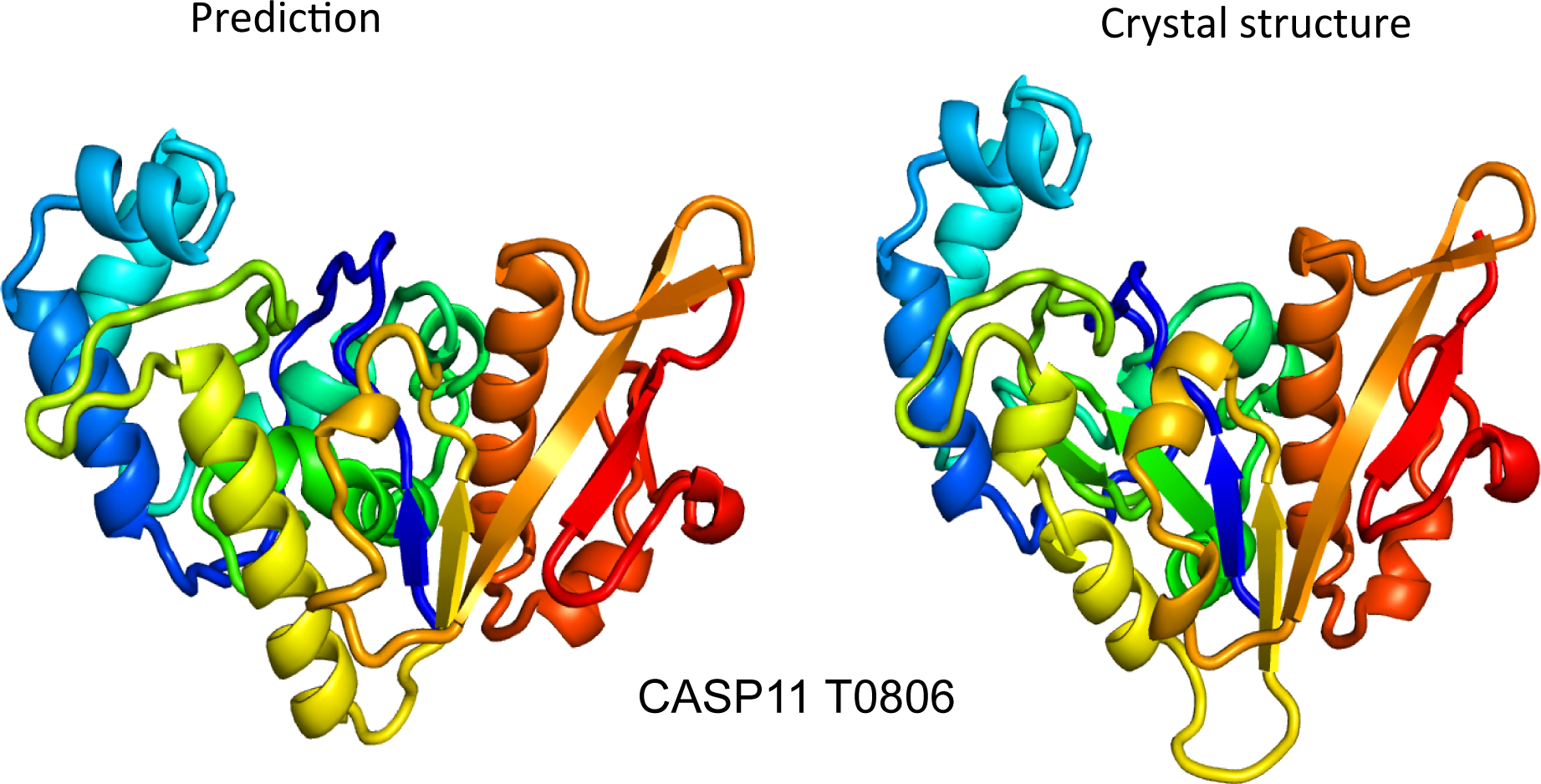
CASP3-11 Results Published in E-Life
In the early 1990s, researchers in the field of protein structure prediction were challenged by the problem of how to impartially judge the accuracy of prediction algorithms. This realization led the protein structure prediction the community to start the Critical Assessment of protein Structure Prediction (CASP), a community-wide, worldwide experiment for protein structure prediction taking…
-
August IPD News Roundup
RESEARCH Following the groundbreaking 2014 Nature paper describing the development of a computational method to design multi-component coassembling protein nanoparticles, comes a publication in Protein Science from Baker lab graduate student Jacob Bale and collaborators. Titled “Structure of a designed tetrahedral protein assembly variant engineered to have improved soluble expression“, the paper reports a variant of a previously low…
-
New Science paper: Designed 2-D protein arrays
A new Science paper is out from IPD faculty Dr. David Baker titled Design of ordered two-dimensional arrays mediated by noncovalent protein-protein interfaces. Read the abstract below and the article at the link: http://www.sciencemag.org/content/348/6241/1365 We describe a general approach to designing two-dimensional (2D) protein arrays mediated by noncovalent protein-protein interfaces. Protein homo-oligomers are placed into one of the…
-
Designer Proteins to Target Cancer Cells
What if scientists could design a completely new protein that is precision-tuned to bind and inhibit cancer-causing proteins in the body? Collaborating scientists at the UW Institute for Protein Design (IPD) and Molecular Engineering and Sciences Institute (MolES) have made this idea a reality with the designed protein BINDI. BINDI (BHRF1-INhibiting Design acting Intracellularly) is a completely novel protein, based…
-
Accurate Design of Co-Assembling Multi-Component Protein Nanomaterials
A new paper is out in the June 5 issue of Nature entitled Accurate design of co-assembling multi-component protein nanomaterials. Scientists at the Institute for Protein Design (IPD), in collaboration with researchers at UCLA and HHMI, have built upon their previous work constructing single-component protein nanocages and can now design and build self-assembling protein nanomaterials made up of…
-
Design of Activated Serine-Containing Catalytic Triads with Atomic-Level Accuracy
Baker lab members published in Nature Chemical Biology a paper entitled “Design of activated serine-containing catalytic triads with atomic-level accuracy“, describing the computational design of proteins with idealized serine-containing catalytic triads which can capture and neutralize organophosphate probes. This work has utility in design of scavengers of environmental toxins. Learn more at this link.
-
Increasing Public Involvement in Structural Biology
Foldit is 5 years old. This publication entitled “Increasing public involvement in structural biology” chronicles the power of engaging the citizen science community on behalf of the computational challenge of protein folding. Learn more at this link.
-
A New Vaccine Design Method To Combat A Dangerous Virus
In a widely cited Nature paper entitled Proof of principle for epitope-focused vaccine design, IPD researchers and collaborators invented a new method to design novel proteins for use as a candidate vaccines to protect against respiratory syncytial virus (RSV), a significant cause of infant mortality. Learn more at this link.
-
Computational Design of a pH Sensitive Antibody Binder
Purification of antibody IgG from crude serum or culture medium is required for virtually all research, diagnostic, and therapeutic antibody applications. Researchers at the Institute for Protein Design (IPD) have used computational methods to design a new protein (called “Fc-Binder”) that is programed to bind to the constant portion of IgG (aka “Fc” region) at basic…
-
Computational Protein Design To Improve Detoxification Rates Of Nerve Agents
V-type nerve agents are among the most toxic compounds known, and are chemically related to pesticides widespread in the environment. Using an integrated approach, described in an ACS Chemical Biology paper entitled Engineering V-type nerve agents detoxifying enzymes using computationally focused libraries, Dr. Izhack Cherny, Dr. Per Greisen, and collaborators increased the rate of nerve…
-
High-Resolution Comparative Modeling with RosettaCM
Researchers in the Baker group describe an improved method for comparative modeling, RosettaCM, which optimizes a physically realistic all-atom energy function over the conformational space defined by homologous structures. Learn more at this link.
-
Computational Design Of A Protein That Binds Polar Surfaces
In a Journal of Molecular Biology publication entitled Computational design of a protein-based enzyme inhibitor, Dr. Erik Procko and collaborators describe the computational design of a protein-based enzyme inhibitor that binds the polar active site of hen egg lysosome (HEL). Computational design of a protein that binds polar surfaces has not been previously accomplished. Learn…
-
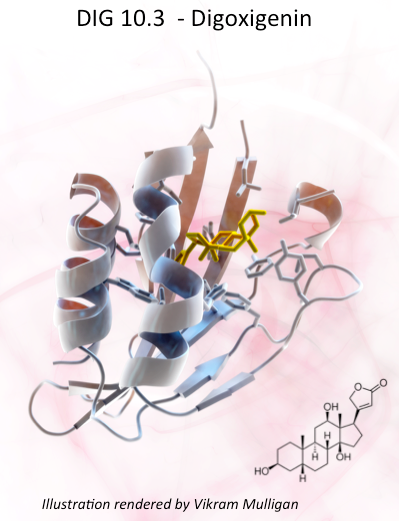
One Small Molecule Binding Protein, One Giant Leap for Protein Design
Reported on-line in Nature (Sept. 4, 2013) researchers at the Institute for Protein Design describe the use of Rosetta computer algorithms to design a protein which binds with high affinity and specificity to a small drug molecule, digoxigenin a dangerous but sometimes life saving cardiac glycoside. Learn more at this link.
-
IPD Researchers Publish New Protocols for Preparing Protein Scaffold Libraries for Functional Site Design
IPD researchers in the Baker group have published new computational protocols for preparing protein scaffold libraries for functional site design. Their paper entitled “A Pareto-optimal refinement method for protein design scaffolds“ improves the search for amino acids with the lowest energy subject to a set of constraints specifying function. Learn more at this link.
-
Centenary Award and Frederick Gowland Hopkins Memorial Lecture
Dr. David Baker, Director of the IPD delivered the Centenary Award and Frederick Gowland Hopkins Memorial Lecture at at the MRC Laboratory of Molecular Biology, Cambridge, UK, on December, 13, 2012. Baker’s lecture entitled “Protein folding, structure prediction and design” can be read at this published link. See: Baker, D. (2014). Protein folding, structure prediction and design.. Biochemical Society…
-
Proteins Made to Order. Researchers at the IPD Design Proteins from Scratch with Predictable Structures
A team from David Baker’s laboratory at the University of Washington in Seattle have described a set of “rules” for the design of proteins from scratch, and have demonstrated the successful design of five new proteins that fold reliably into predicted conformations. Their work was published Nature. Learn more at this link.
-
Computer Designed Proteins Programmed to Disarm a Variety of Flu Viruses
As reported in Nature Biotechnology, David Baker and scientists at the IPD published exciting new methods to improve the potency and breadth of computer-designed protein inhibitors of influenza. Learn more at this link.
-
Computational Design of Self-Assembling Protein Nanomaterials with Atomic Level Accuracy
IPD researchers in the Baker group have published in Science a paper entitled “Computational design of self-assembling protein nanomaterials with atomic level accuracy.” They describe a general computational method for designing proteins that self-assemble to a desired symmetric architecture. Protein building blocks are docked together symmetrically to identify complementary packing arrangements, and low-energy protein-protein interfaces are then designed…
