Citizen scientists can now use Foldit to successfully design synthetic proteins. The initial results of this unique collaboration appear today in Nature.
Brian Koepnick, a recent PhD graduate in the Baker lab, led a team that worked on Foldit behind the scenes, introducing new features into the game that they believed would help players home in on better folded structures.
Players were provided with a poly-isoleucine backbone in a fully extended conformation (60-100 residues in length). They had seven days to fold the backbone into a compact structure and assign a sequence specifying this new structure.
Scores in Foldit are calculated using Rosetta. By competing with one another to reach the highest score, Foldit players arrive at virtual proteins with extremely low energies (a high Foldit score corresponds to low protein energy). But since energy alone is not enough for protein design, the Foldit team made adjustments to the Foldit score function. These included requiring the presence of a hydrophobic core, limiting the placement of glycine and alanine, and other side-chain specific terms.
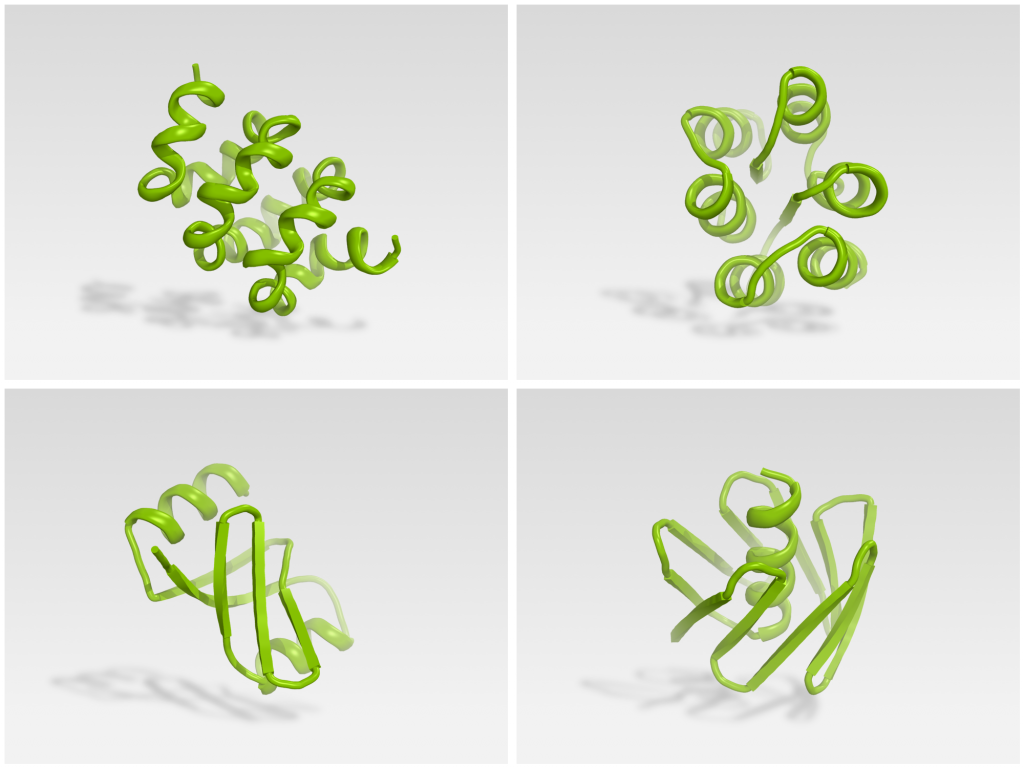
The team experimentally tested 146 Foldit designs. 56 were found to be stable monomers when expressed in E coli. X-ray crystallography and NMR were used to determine the structure of four Foldit designs, which agreed strongly with their design model.
Every step of the way, the team relied on the work of Foldit players to expose problems with the score function. Foldit players are excellent at exploring new kinds of protein folds. For this reason, Foldit players are incredibly helpful for identifying unanticipated weaknesses in Rosetta, and ultimately can improve our understanding of protein folding.
Now that Foldit players can accurately design high-quality proteins from scratch, we can start to challenge Foldit players with more applied protein design problems. Foldit players can now help to design proteins that can assemble into multi-component structures, or that can bind to biological targets as potent medicines, or that can degrade toxic chemicals.
Because Foldit depends on the cooperation and competition of its player community, our scientific ability grows rapidly with the number of Foldit players. We look forward to expanding the Foldit community and recruiting more creative and curious Foldit players!
Read the full manuscript: https://rdcu.be/bFE7R PDF