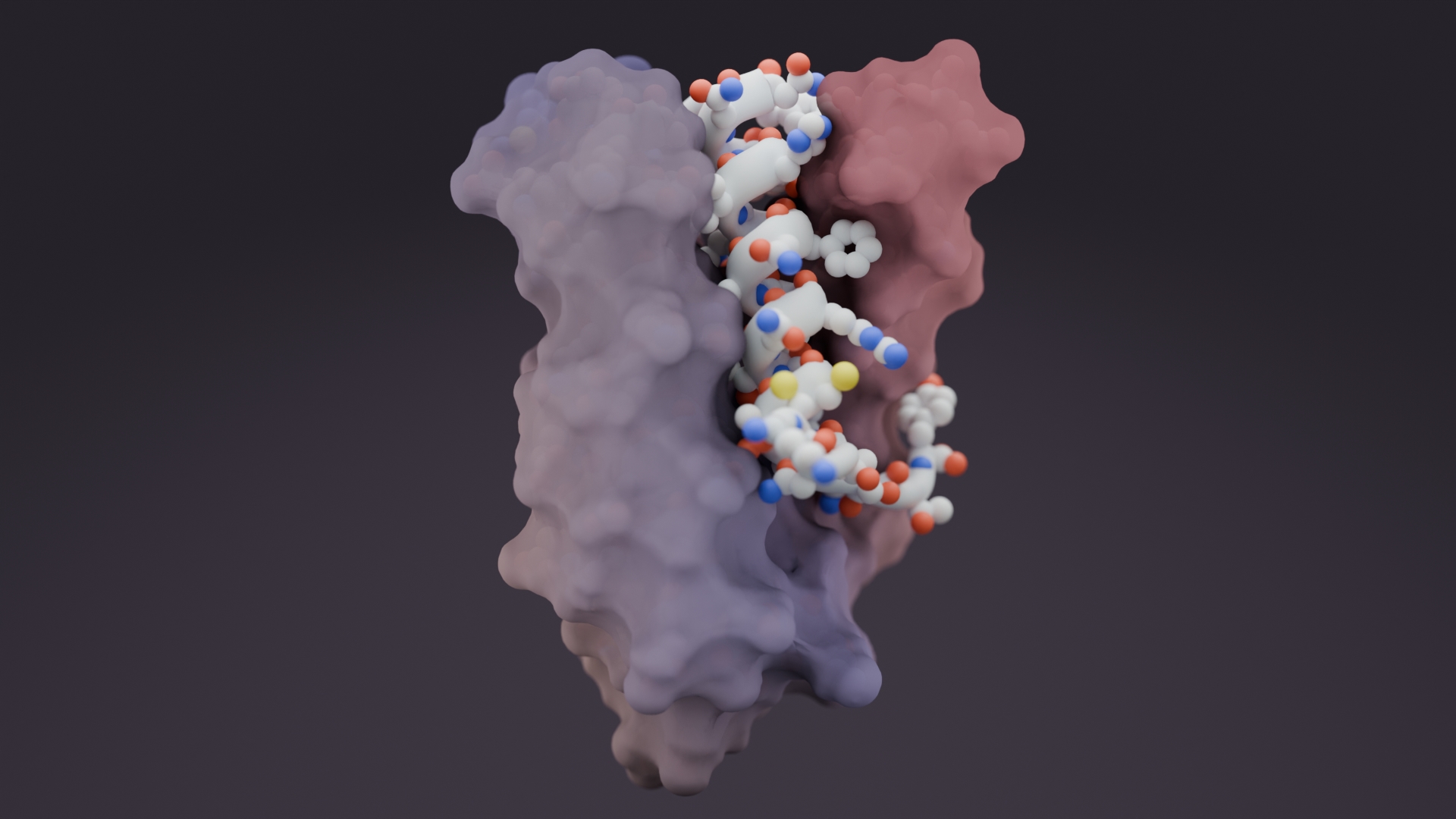
A team including scientists from the Baker Lab has published in Nature this week on the development and initial applications of RFdiffusion. This guided diffusion model for generating new proteins outperforms prior protein design methods across a broad range of problems.
From Nature:
“OK. Here we go.” David Juergens, a computational chemist at the University of Washington (UW) in Seattle, is about to design a protein that, in 3-billion-plus years of tinkering, evolution has never produced.
On a video call, Juergens opens a cloud-based version of an artificial intelligence (AI) tool he helped to develop, called RFdiffusion. This neural network, and others like it, are helping to bring the creation of custom proteins — until recently a highly technical and often unsuccessful pursuit — to mainstream science.
These proteins could form the basis for vaccines, therapeutics and biomaterials. “It’s been a completely transformative moment,” says Gevorg Grigoryan, the co-founder and chief technical officer of Generate Biomedicines in Somerville, Massachusetts, a biotechnology company applying protein design to drug development.
The tools are inspired by AI software that synthesizes realistic images, such as the Midjourney software that, this year, was famously used to produce a viral image of Pope Francis wearing a designer white puffer jacket. A similar conceptual approach, researchers have found, can churn out realistic protein shapes to criteria that designers specify — meaning, for instance, that it’s possible to speedily draw up new proteins that should bind tightly to another biomolecule. And early experiments show that when researchers manufacture these proteins, a useful fraction do perform as the software suggests.
The tools have revolutionized the process of designing proteins in the past year, researchers say. “It is an explosion in capabilities,” says Mohammed AlQuraishi, a computational biologist at Columbia University in New York City, whose team has developed one such tool for protein design. “You can now create designs that have sought-after qualities.”
“You’re building a protein structure customized for a problem,” says David Baker, a computational biophysicist at UW whose group, which includes Juergens, developed RFdiffusion. The team released the software in March 2023, and a paper describing the neural network appears this week in Nature1. (A preprint version was released in late 2022, at around the same time that several other teams, including AlQuraishi’s2 and Grigoryan’s3, reported similar neural networks).
For the first time, protein designers now have the kinds of reproducible and robust tools around which a new industry can be created, Grigoryan adds. “The next challenge becomes, what do you do with it?”
Read the full story at nature.com