In a recent preprint, we describe deep-learning breakthroughs that significantly expand the types of proteins and protein assemblies that can be modeled and designed using computers.
This work updates two of our most powerful software tools, RoseTTAFold and RFdiffusion, by training them to operate on additional types of molecules beyond just proteins. Through these efforts, we aim to both increase our understanding of life and accelerate the design of new proteins with advanced functions.
This project was led by postdoctoral scholar Jue Wang and graduate students Rohith Krishna and Woody Ahern, who are all members of the Baker Lab at the Institute for Protein Design at the University of Washington School of Medicine.



RoseTTAFold All-Atom models more of life’s molecules
The team began by making changes to RoseTTAFold, which, like AlphaFold, is a highly accurate deep-learning program for modeling protein structures. These programs were developed to model only biomolecules made entirely of amino acids. The new upgrades now allow RoseTTAFold All-Atom to model full biological assemblies that contain many different types of molecules, including proteins, DNA, RNA, small molecules, metals, and other bonded atoms, including covalent modifications of proteins.
This is important for understanding biology because proteins rarely function on their own and instead must interact with other non-protein compounds. RoseTTAFold All-Atom may also benefit drug discovery research by allowing scientists to model how proteins and small-molecule drugs interact.
“We’ve expanded our modeling capabilities beyond amino acids, which should bring clarity to new aspects of molecular biology. It’s a bit like switching from black and white to a color TV,” said Krishna.
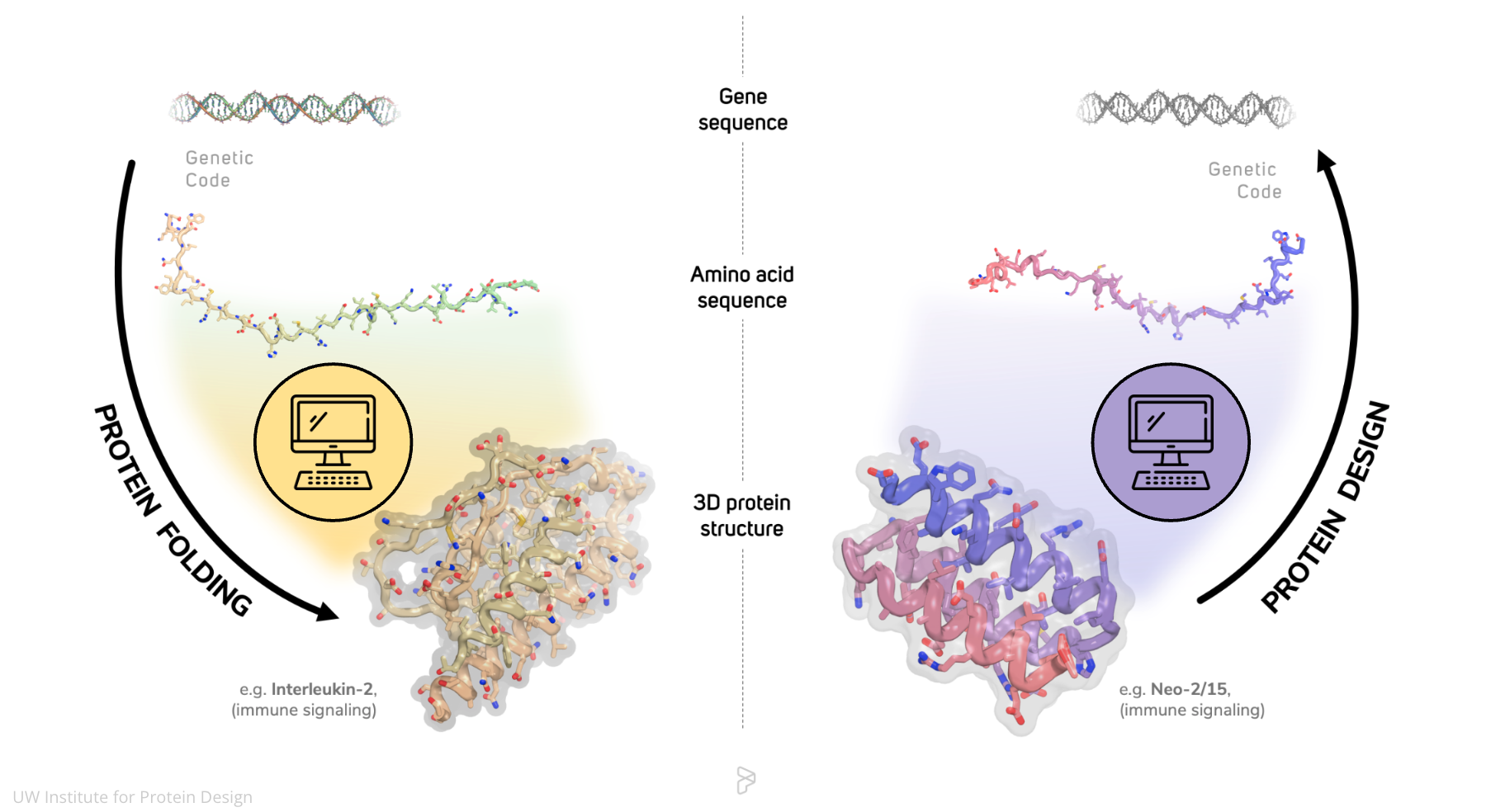
RFdiffusion All-Atom generates proteins with advanced functions
This upgrade to RoseTTAFold allowed the team to create a new version of RFdiffusion, which is our powerful deep-learning program that can generate new protein structures in a manner similar to how DALL-E or Midjourney generate art.
Through extensive laboratory testing, we confirmed that the updated design tool, dubbed RFdiffusion All-Atom, yields proteins with advanced functions, including the ability to bind to specific small molecules like heme or the heart disease drug digoxigenin.
“Our improved protein design software will allow us and others to create an even wider range of versatile and functional molecules,” said Ahern. “I can’t wait to see all the ways scientists will use these tools.”
The ability to create proteins that are programmed to bind to specific target molecules sets the stage for the development of new diagnostics, gene-editing technologies, enzymes, and more. The ability to model full biomolecular systems should considerably extend the power of deep learning tools for protein structure prediction, leading to a richer understanding of biology.
This research was posted on bioRvix on Oct. 9 and has not yet been peer-reviewed. The initial development and applications of RoseTTAFold were reported in Science in 2021. The initial development and applications of RFdiffusion were reported in Nature in 2022.
Funding
This work was supported by The Audacious Project, Howard Hughes Medical Institute, Bill and Melinda Gates Foundation (OPP1156262), Open Philanthropy (INV-010680), Schmidt Futures, Helmsley Charitable Trust (2019PG-T1D026), Juvenile Diabetes Research Foundation (2-SRA-2018-605-Q-R), Microsoft, Amgen, Seoul National University, University of Sheffield, Washington State General Operating Fund, Defense Threat Reduction Agency (HDTRA1-19-1-0003), Department of Health and Human Services (75N93022C00036), National Energy Research Scientific Computing Center (BER-ERCAP0022018), National Institutes of Health, European Research Council, Royal Society University Research Fellowship (URF\R1\191548), and the Human Frontiers Science Program (RGP0061/2019).





