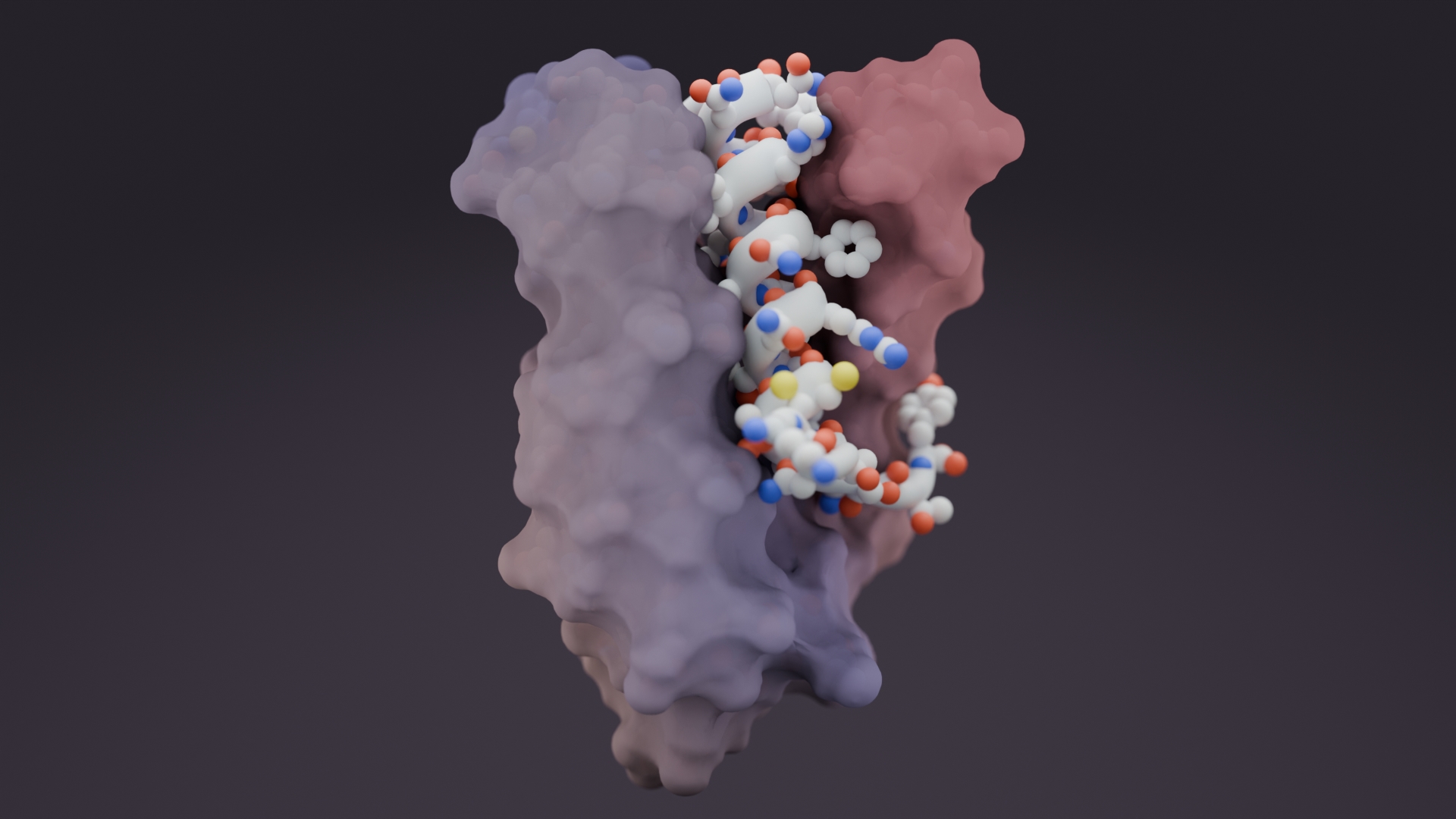
In nature, many proteins fold into stable shapes. But intrinsically disordered proteins (IDPs) and regions (IDRs) — which make up nearly half of the human proteome — lack consistent structures. These molecules drive key cellular signaling, stress responses, and disease progression yet have long been challenging to study or target with new medicines due to their high conformational flexibility.
In two new studies, researchers in the Baker Lab show that generative AI can be used to create proteins that bind highly flexible proteins and peptides with atomic precision, tackling targets long considered impossible to drug. The initial proteins produced with these methods blocked pain signaling, dismantled toxic aggregates, and disabled prion seeds in cell-based tests.
Publications
Design of intrinsically disordered region binding proteins
Authors: Kejia Wu, Hanlun Jiang, Derrick R. Hicks, Caixuan Liu, Edin Muratspahić, Theresa A. Ramelot, Yuexuan Liu, Kerrie McNally, Sebastian Kenny, Andrei Mihut, Amit Gaur, Brian Coventry, Wei Chen, Asim K. Bera, Alex Kang, Stacey Gerben, Mila Ya-Lan Lamb, Analisa Murray, Xinting Li, Madison A. Kennedy, Wei Yang, Zihao Song, Gudrun Schober, Stuart M. Brierley, John O’Neill, Michael H. Gelb, Gaetano T. Montelione, Emmanuel Derivery, David Baker
Diffusing protein binders to intrinsically disordered proteins
Authors: Caixuan Liu, Kejia Wu, Hojun Choi, Hannah Han, Xulie Zhang, Joseph L. Watson, Sara Shijo, Asim K. Bera, Alex Kang, Evans Brackenbrough, Brian Coventry, Derrick R. Hick, Andrew N. Hoofnagle, Ping Zhu, Xingting Li, Justin Decarreau, Stacey R. Gerben, Wei Yang, Xinru Wang, Mila Lamp, Analisa Murray, Magnus Bauer, David Baker
Lead Authors




Coverage
David Baker’s lab uses AI to help catch nature’s squirmiest proteins — Endpoints News
Undruggable ‘disordered’ proteins become druggable with new AI techniques — STAT News
UW scientists use AI to crack ‘undruggable’ proteins, opening door to new treatments — GeekWire
Undruggable No More: AI Hits Disordered Proteins, Unlocks Therapy Targets — GEN
Introducing ‘logos’
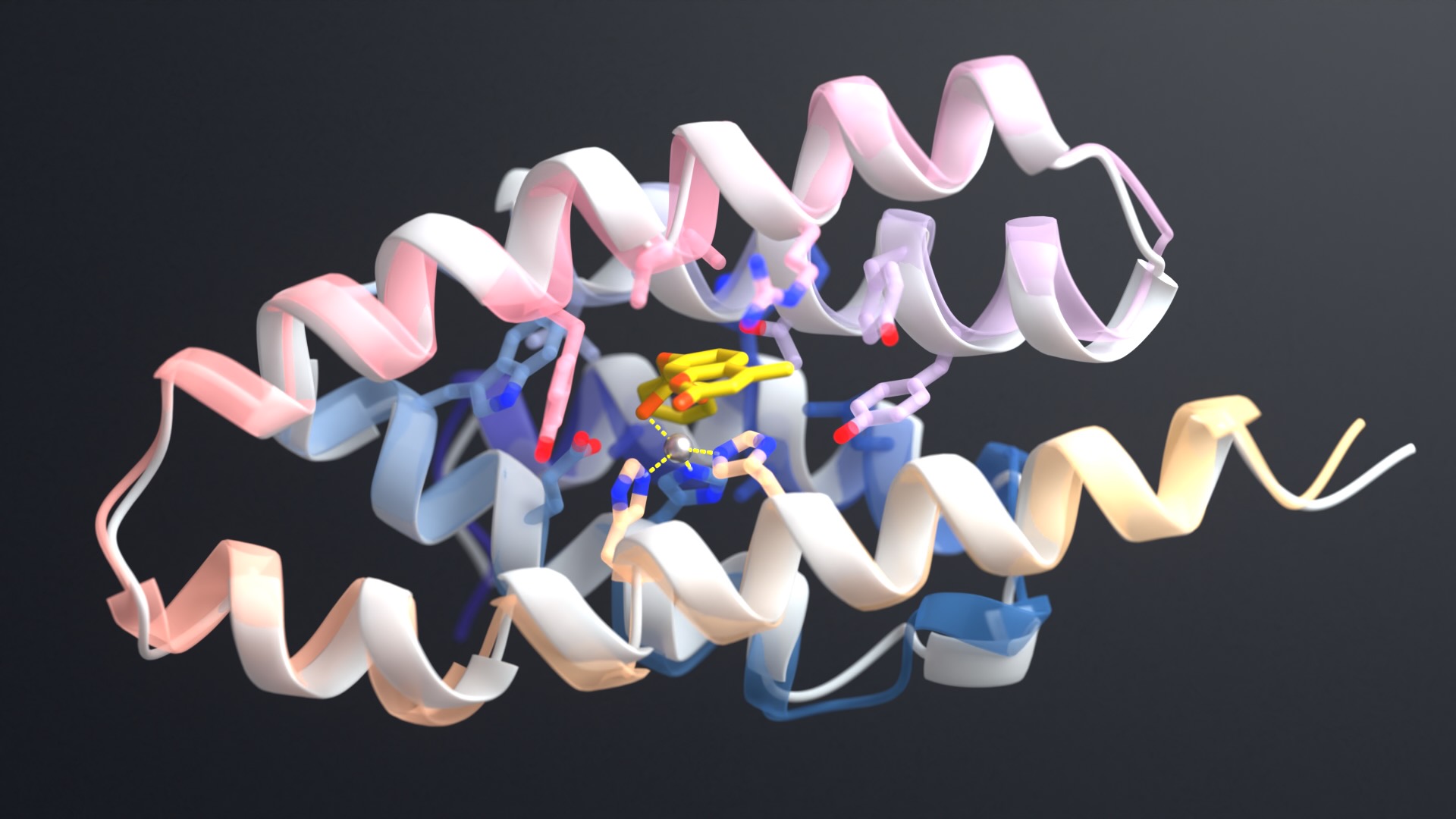
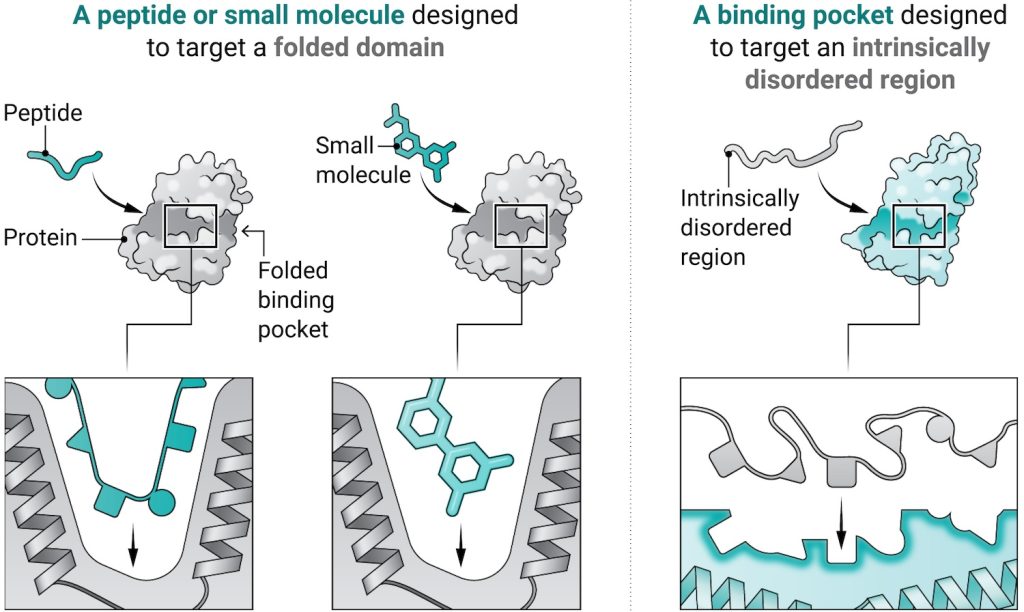
One study published today in Science introduces a design strategy called ‘logos’ for building binders to virtually any disordered protein or peptide target. By assembling binding proteins from a library of 1,000 pre-made parts, the team created tight binders for 39 of 43 targets tested. One such binder, aimed at the opioid peptide dynorphin, blocked pain signaling inside lab-grown human cells.
“To show our approach is truly general, we even built binders for peptides encoding random English words. A thousand prefabricated pockets allow for trillions of combinations.”
Kejia Wu, PhD
A diffusion approach
In a complementary study in Nature, we applied RFdiffusion to generate proteins that wrap around flexible targets. They team produced high-affinity binders (3–100 nM) for amylin, C-peptide, VP48, G3BP1, the IL-2 receptor γ-chain, and the pathogenic prion core. The amylin binders dissolved amyloid fibrils linked to type 2 diabetes in lab tests.
“We produced binders that are highly specific and match nature’s strongest interactions, reaching nanomolar to picomolar affinities — a critical capability that wasn’t possible before.”
Caixuan Liu, PhD
Complementary strategies
“The RFdiffusion-based method excels at designing binders to targets with some helical and strand secondary structure, while the logos method works best for targets lacking regular secondary structure.”
David Baker, PhD
Since many disease drivers contain disordered segments, these new design strategies could spark a wave of new therapeutics and diagnostics. The protein design software that enabled this project is freely accessible to all researchers.
This research was supported by several grants and philanthropic donors. All funders are listed in the manuscripts.