Two studies published this week by IPD researchers highlight a trend in AI-accelerated protein research: entire multi-protein complexes can now be generated from scratch using life’s most functional building blocks. This leap forward in biotechnology promises to transform how medicines, materials, and molecular tools are made.
Led by the Baker Lab and King Lab and published in Nature Materials, the new papers introduce complementary approaches for designing custom protein systems with unprecedented control over size, shape, and function. As in nature, these nanoscale architectures assemble themselves through chemical forces. But unlike natural protein assemblies, they are programmed on computers with precise molecular features in mind.
Publications
Bond-centric modular design of protein assemblies
Published in: Nature Materials
Authors: Shunzhi Wang, Andrew Favor, Ryan D. Kibler, Joshua M. Lubner, Andrew J. Borst, Nicolas Coudray, Rachel L. Redler, Huat Thart Chiang, William Sheffler, Yang Hsia, Neville P. Bethel, Zhe Li, Damian C. Ekiert, Gira Bhabha, Lilo D. Pozzo, David Baker
Computational design of bifaceted protein nanomaterials
Published in: Nature Materials
Authors: Sanela Rankovic, Kenneth D. Carr, Justin Decarreau, Rebecca Skotheim, Ryan D. Kibler, Sebastian Ols, Sangmin Lee, Jung-Ho Chun, Marti R. Tooley, Justas Dauparas, Helen E. Eisenach, Matthias Glögl, Connor Weidle, Andrew J. Borst, David Baker, Neil P. King
Lead Authors


Anisotropic Assemblies

Like the Roman deity Janus, each bifaceted nanoparticle has two distinct faces. This enables the particles to present different functional features at precisely defined distances. Image made with ChatGPT by Ian C. Haydon, UW Institute for Protein Design.
One study, led by Sanela Rankovic, PhD, introduces a method for designing protein nanoparticles with two unique faces. These Janus-like structures can bring together distinct biological targets at controlled distances, offering a powerful new way to organize living cells, signaling molecules, and other tiny objects.
Lab tests reveal that these assemblies can be used to bridge populations of receptor-coated microparticles as intended, a demonstration of their potential for cellular engineering.
“I’m particularly excited about the application of bifaceted structures as next-generation vaccine platforms and in cancer immunotherapy. Depending on how they are functionalized, these structures could enable the co-delivery of antigens and immunomodulatory molecules to create programmed interactions between target cells, such as T cells and cancer cells, with precise control over spacing.”
Sanela Rankovic, PhD
Modular Assemblies
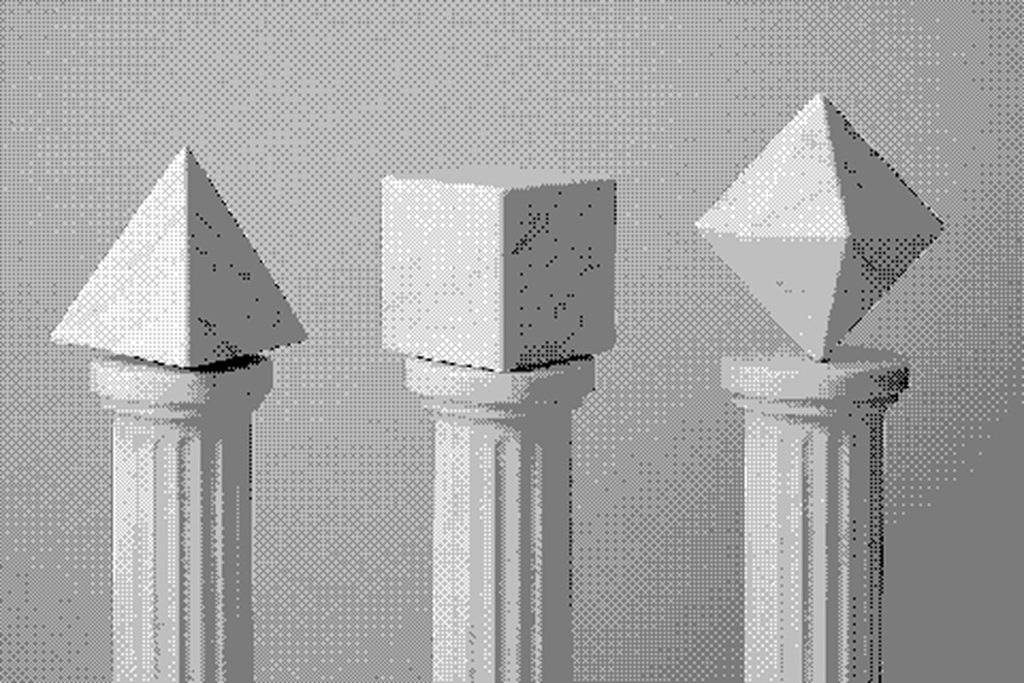
Separately, Shunzhi Wang, PhD, and colleagues developed a generalizable protein design strategy inspired by chemistry’s economy of parts. Just as atoms bond to each other in regular ways to form larger molecules, their approach created a small set of reusable protein building blocks that enable the assembly of polyhedral cages, two-dimensional lattices, and three-dimensional crystalline networks.
Some of these systems were shown to be reconfigurable, dynamically switching between architectures under different laboratory conditions.
“Watching a pellet of 2D protein sheets dissolve and, with one extra building block, re-form as tiny cages felt like a magic trick. Our work shows how new protein design approaches enable a tiny protein ‘alphabet’ to spell out an awful lot of molecular ‘sentences.’”
Shunzhi Wang, PhD
The ability to produce reusable protein parts shifts protein design closer to the long-standing goal of being able to generate custom nanostructures on demand, and with a key advantage: proteins offer the greatest functional diversity of any molecules in the known universe.
An Inverted Strategy
Both papers demonstrate a reversal of how synthetic protein complexes were built in the past.
“Traditionally, new protein nanomaterials were built by docking symmetrical protein units into target architectures and then designing interfaces to hold them together. These new methods reverse that process: the interfaces between building blocks are arranged in space first, and then bespoke protein subunits are generated to scaffold them in place. The result is a versatile strategy for building complex multi-component systems that nature never evolved.”
Neil King, PhD, associate professor of biochemistry
Funding
This work was supported by the Bill & Melinda Gates Foundation, National Institutes of Health, Howard Hughes Medical Institute, Burroughs Wellcome Fund, Defense Threat Reduction Agency, and several other organizations. All funders are listed in the manuscripts.