Our Research
By unlocking the power of proteins, we are setting the stage for a new wave of medicines and materials with previously unimagined functions — tapping into the full efficiency, scalability, and sustainability of biology.

Protein Structure Prediction
Our scientists are building deep learning tools that quickly and accurately model the three-dimensional structures of proteins from sequence data alone. Our latest tool RoseTTAFold is available for free public use.
Protein & Small Molecule Binding
We create methods for generating high-affinity protein binding and applying this to targets of medical interest. We are also designing proteins that bind to small molecules. This research provides insights into the molecular interactions that underlie most cellular processes.
Self-Assembling Nanomaterials
Our researchers are developing methods to create new self-assembling nanostructures and using these approaches to develop advanced vaccines and drug delivery vehicles. Our first approved medicine, a vaccine for COVID-19, employs our computationally designed protein nanoparticle technology.
New Protein Scaffolds
In the past, almost all protein design efforts have modified proteins from nature. We are developing methods for designing a wide range of exceptionally stable protein folds from scratch with tunable geometries for specific applications.
Enzyme Design
Enzymes catalyze chemical reactions that are essential for life. We are developing methods to design new enzymes for chemical transformations not found in nature. This research helps reveal more about the chemical foundations of life.
Protein Structure Determination
Our researchers are developing methods for solving protein structures using limited experimental data. These tools are in use in research laboratories around the world.
Applications
From detecting fentanyl to blocking the virus that causes COVID-19, our scientists are pushing the limits of what proteins can do to help meet the challenges of the modern world.

Ultrapotent Vaccines
In 2019, we designed our first synthetic nanoparticle vaccine targeting the respiratory virus RSV, a particularly lethal virus for infants and the elderly. Icosavax, an IPD spinout, is now developing this and other vaccine candidates.
We applied this same platform technology to create SKYCovione, a fully-approved COVID-19 vaccine that contains 60 copies of the SARS-CoV-2 Spike protein receptor binding domain. This potent vaccine does not require freezing, a property that should make it easier to reach communities in need.
Efforts are now underway to develop vaccines that protect against a wide range of related influenza and coronaviruses. Such vaccines would provide protection even in the face of new viral variants.
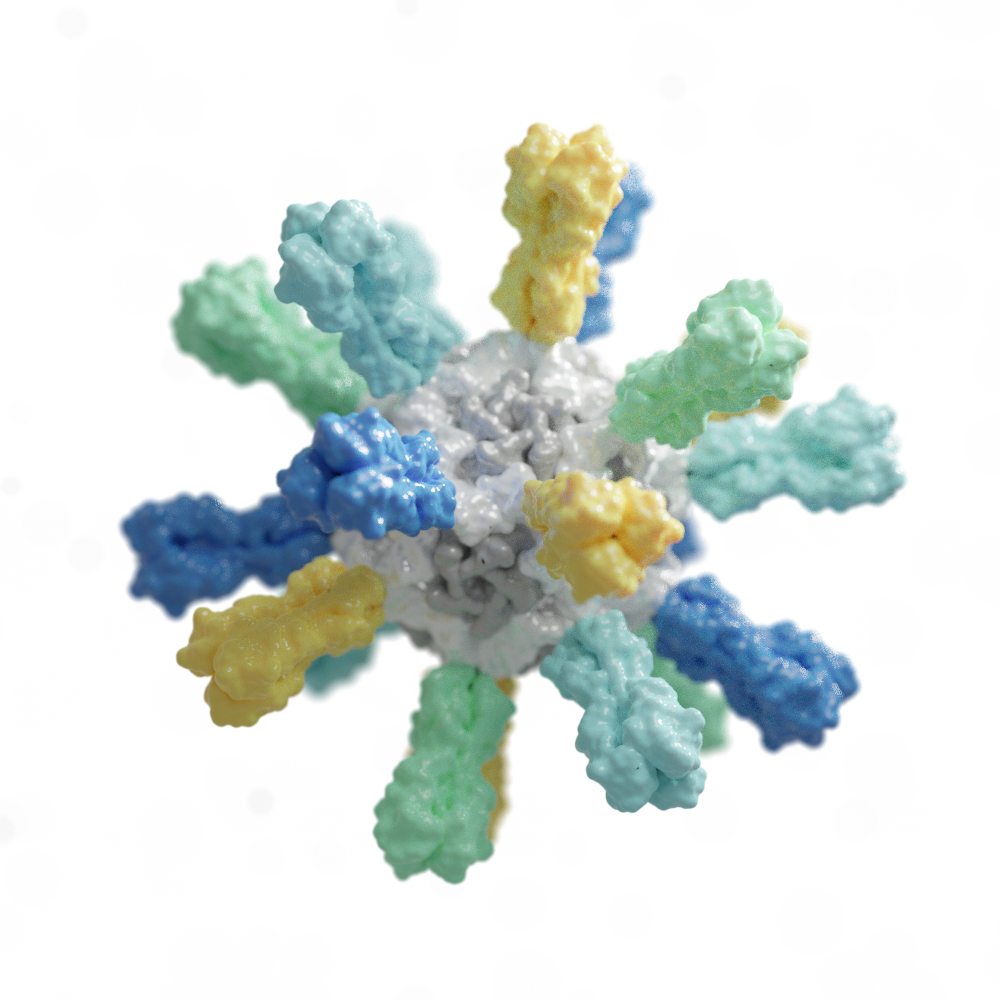
Rapid Antivirals
In 2011 we designed proteins that target the influenza virus. These durable antivirals protect rodents from exposure to lethal amounts of the virus and can be used as diagnostic reagents in low-cost diagnostic test strips, providing improved performance as an influenza assay compared to a traditional antibody-based capture system.
We used this same technology to create potent antivirals for the COVID-19 virus and have adapted these new proteins into sensitive biosensors.
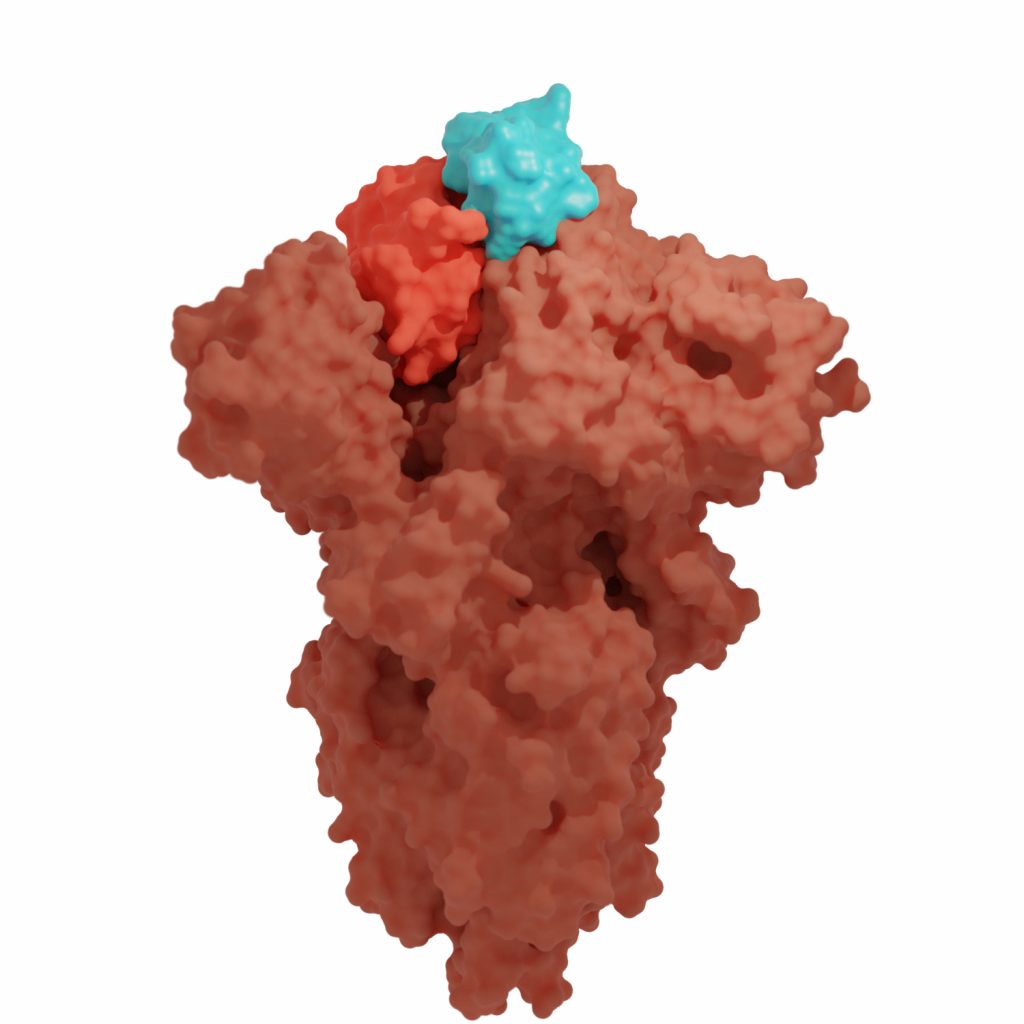
Cancer Immunotherapy
In 2019 we created compact proteins that stimulate the same receptors as IL-2, a powerful immunotherapeutic drug, while avoiding unwanted off-target receptor interactions. These synthetic proteins shrink tumors in mice and may become the basis for a safer type of cancer therapy.
This technology is now being developed by our spinout company Neoleukin Therapeutics.
Celiac Disease
In 2011 a team of UW undergraduates working out of the IPD won the grand prize at the annual iGEM competition. They sought to develop a cure for celiac disease and used computational design to re-engineer a natural enzyme to break down gluten in the harsh acidic conditions of the stomach.
In 2015 our team published the most advanced version of the enzyme, dubbed Kuma030, which cleaves gluten molecules at sites that are known to cause an immune reaction in those with celiac disease. The enzyme was further developed by our spinout company PvP Biologics, which has been acquired by Takeda.
New Nanomaterials
Our researchers are inventing new ways to pattern matter at the nanoscale. Drawing inspiration from natural protein materials such as wool, silk, and even bone, we seek to create a new generation of biomaterials with unrivaled physical properties. These novel materials may replace many polluting petrochemicals, or give rise to new substances with unimagined functions.

