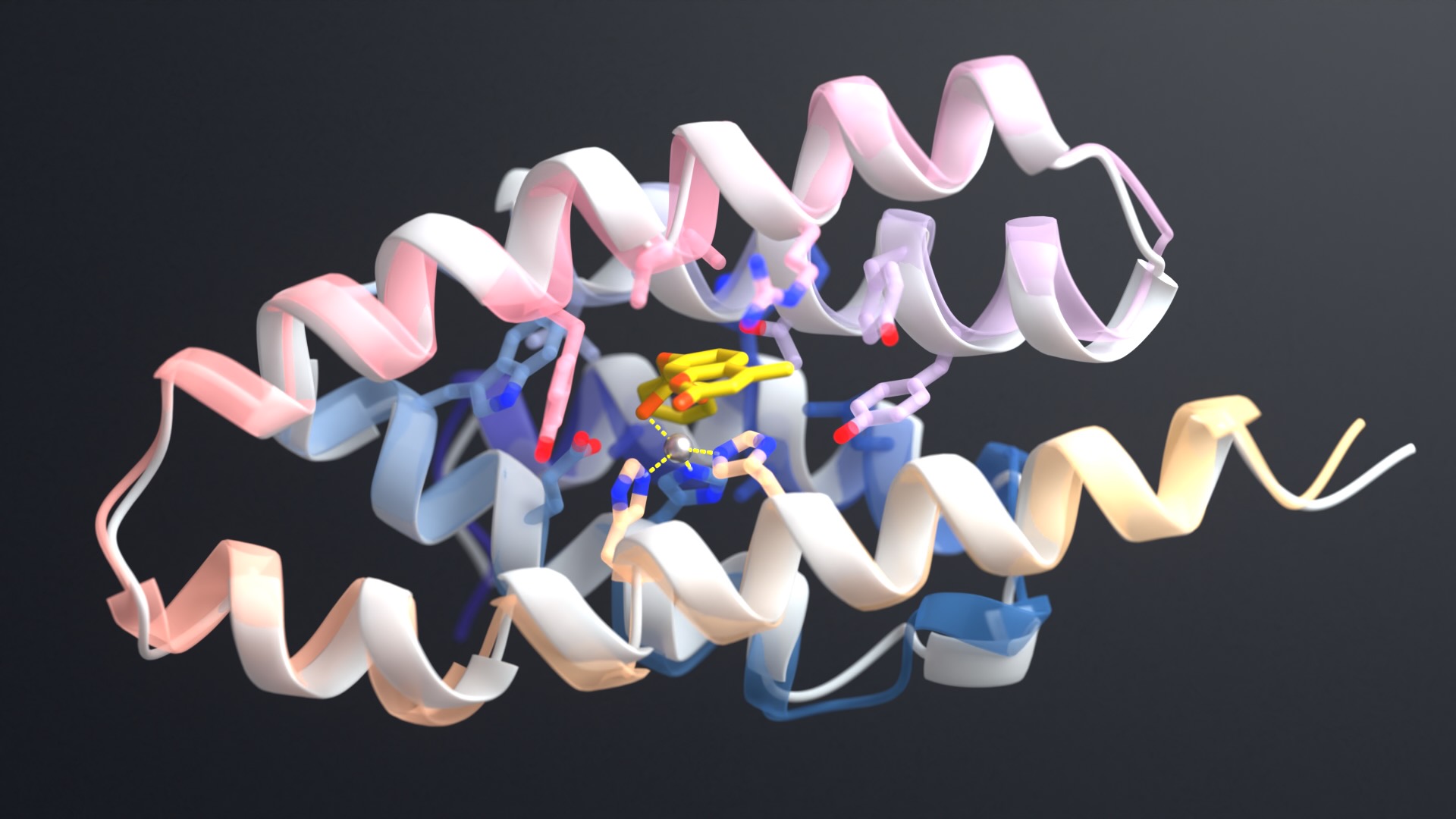
Update (July 2023): Our manuscript on the development of RFdiffusion has been published in Nature.
A team led by scientists from the Baker Lab has created a powerful new way of designing proteins that combines structure prediction networks and generative diffusion models.
The team demonstrated extremely high computational success using the new method and experimentally tested hundreds of A.I.-generated proteins, finding that many may be useful as medications, vaccines, or even new nanomaterials.
Originally appearing as a preprint, this research is now available in Nature. Additional applications of RFdiffusion are also described in a companion preprint.
Drawing inspiration from DALL-E
The software tool DALL-E produces high-quality images that have never existed before using a machine-learning tool called a diffusion model, which is an algorithm that specializes in adding and removing noise.
Diffusion models for image generation begin with grainy bits of static and gradually remove noise until a clear picture is formed. Additional pieces of software guide this denoising process so that the new images end up matching what was asked for.
Drawing inspiration from this work, we have developed a guided diffusion model for generating new protein molecules called RFdiffusion. With prior protein design software, tens of thousands of designed molecules may have to be tested before finding a single one that performs as intended. Using the new method, the team had to test as little as one per design challenge.
RFdiffusion was developed by a team of computational biologists from UW Medicine, Columbia University, and MIT. The project was led by Joseph Watson, David Juergens, Nathaniel Bennett, Brian Trippe, Jason Yim, Helen Eisenach, Woody Ahern.
A new state-of-the-art
RFdiffusion outperforms existing protein design methods across a broad range of problems. These include topology-constrained protein monomer design, protein binder design, symmetric oligomer design, enzyme active site scaffolding, and symmetric motif scaffolding for therapeutic and metal-binding protein design.
So far, we have used RFdiffusion to generate ultra-high affinity binders and a series of novel symmetric assemblies experimentally confirmed by electron microscopy.
“These works reveal just how powerful diffusion models can be for protein design,” says Watson. “It’s extremely exciting,” added Juergens, “and it’s really just the beginning.