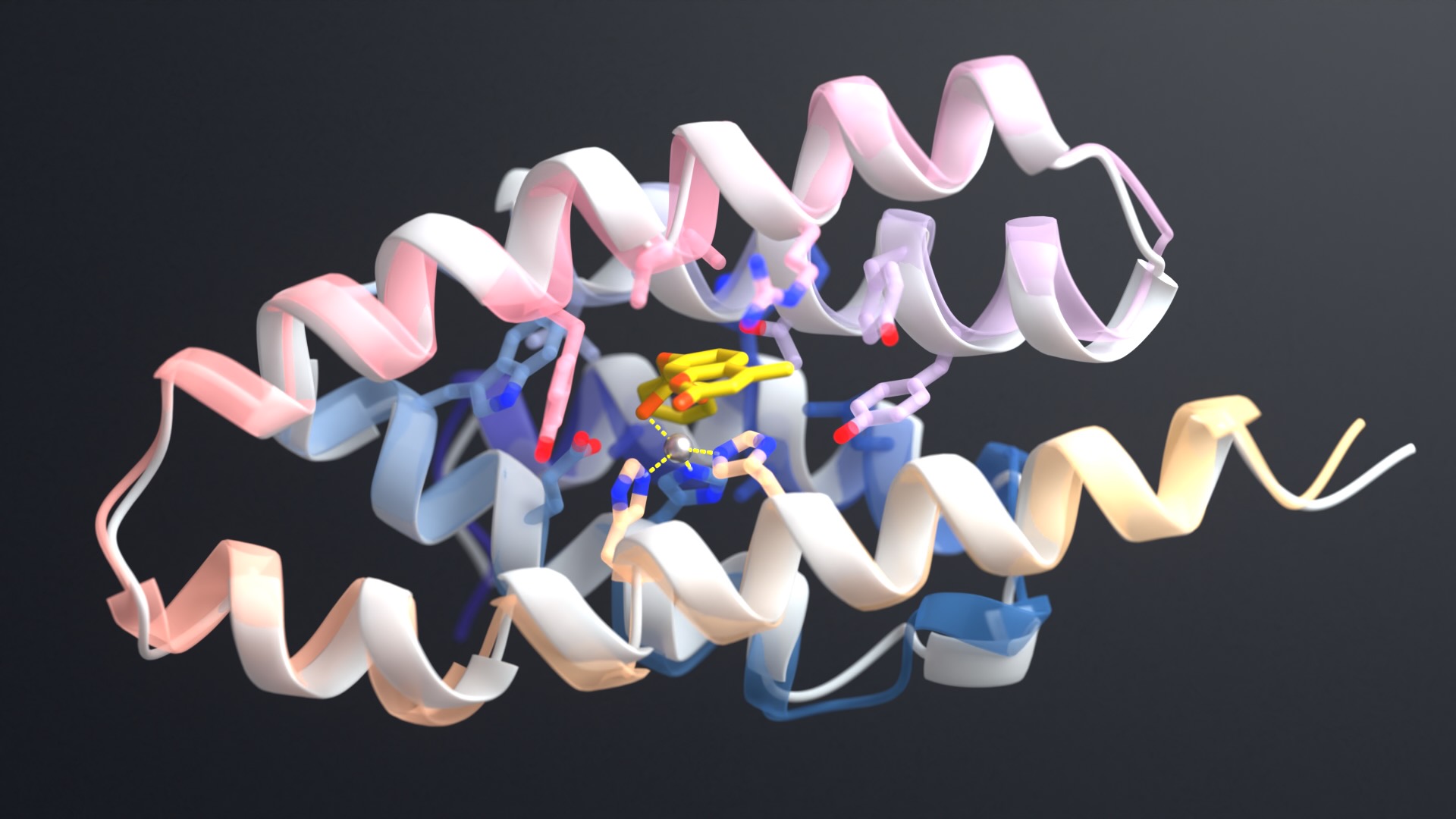
A new study led by the Baker Lab shows for the first time that functional, full-length antibodies can be developed entirely on computers — no animal immunizations or extensive screening required. This breakthrough could reshape the $200 billion antibody drug industry and open pathways to treatments for diseases currently considered undruggable.
The work builds on a preprint we posted in March 2024 showing AI could generate functional nanobody fragments. Today’s publication marks a significant advance: it is now possible to design complete antibody variable regions containing both heavy and light chains — the molecular architectures used in most current antibody drugs.
“AI antibody design will transform the biotechnology and pharmaceutical industries, enabling precise targeting and simpler drug development.”
David Baker, PhD, senior author and director of the Institute for Protein Design
We’re sharing the antibody design software RFantibody freely for academic, personal, and commercial use, enabling laboratories worldwide to apply this technology. The code is available on GitHub.
Publications
Atomically accurate de novo design of antibodies with RFdiffusion
Authors: Nathaniel R. Bennett, Joseph L. Watson, Robert J. Ragotte, Andrew J. Borst, DéJenaé L. See, Connor Weidle, Riti Biswas, Yutong Yu, Ellen L. Shrock, Russell Ault, Philip J. Y. Leung, Buwei Huang, Inna Goreshnik, John Tam, Kenneth D. Carr, Benedikt Singer, Cameron Criswell, Basile I. M. Wicky, Dionne Vafeados, Mariana Garcia Sanchez, Ho Min Kim, Susana Vázquez Torres, Sidney Chan, Shirley M. Sun, Timothy T. Spear, Yi Sun, Keelan O’Reilly, John M. Maris, Nikolaos G. Sgourakis, Roman A. Melnyk, Chang C. Liu, David Baker
Lead Authors







Coverage
AI-designed antibodies promise big boost to drug development — Financial Times
‘Not the end of the story:’ Nobel laureate David Baker on what’s next for de novo design after RFantibody lands in Nature — Endpoints News
AI-Designed Antibodies Achieve Atomic Precision to Enhance Drug Discovery — GEN
Academic AIs make inroads in protein binder design — C&EN
AI tuned for antibody design
Antibodies are proteins that recognize and bind to specific sites on target molecules, triggering protective responses in the body. The pharmaceutical industry has turned them into blockbuster drugs for cancer, autoimmune disease, and other conditions.
Discovering new therapeutic antibodies typically requires months of animal immunization or screening millions of randomly generated molecules. These processes are expensive, time-consuming, and often fail to find useful antibodies.
The team trained a version of RFdiffusion — a generative AI tool that works like DALL-E but for molecular structures — to design antibody binding regions. The model learned to create the molecular “fingertips” that grip onto targets with near-atomic precision, matching researcher specifications.
“When we first announced RFdiffusion in 2023, we showed that generative AI is a powerful tool for creating new molecules. This new work, where we extended these methods to antibody design, demonstrates that rational design of antibodies is now possible, and takes a big step towards delivering better therapeutics for patients.”
Joe Watson, co-lead author
The team combined computational design with high-throughput screening and laboratory evolution to identify and optimize functional candidates.
While deep learning has been used before to optimize existing antibodies, this marks the first time AI models can produce new antibodies for targets that lack known binding partners.
“Many diseases have no approved medicines because they are triggered by molecules in the body with vulnerable sites that are difficult to target with traditional small-molecule drugs. Computer-generated antibodies could target these sites directly.”
Nate Bennett, PhD co-lead author
Blocking viruses, toxins and more
The team tested the antibody design method against several challenging targets: toxins from disease-causing bacteria, viral proteins from influenza and coronavirus, and notoriously difficult cancer targets. In most cases, they identified AI-generated antibodies that functioned as intended.
For antibodies targeting Clostridium difficile toxin, the team achieved mid-nanomolar binding affinities — comparable to therapeutic antibodies in clinical use. When converted to full IgG format (the structure used in drugs like Humira and Keytruda), the molecules retained their potency.
Using electron microscopy, the team examined five AI-designed antibodies bound to their targets. Four matched the computer predictions perfectly, gripping their targets exactly where and how the AI intended.
“Our testing confirmed that many of the computer designs were essentially perfect. Those antibodies fold exactly as predicted and bind their targets exactly where intended. Seeing where the pipeline could be refined will help further improve success rates moving forward.”
Andrew Borst, PhD co-lead author and head of electron microscopy at the Institute for Protein Design
Transforming drug development
The team believes computational design could compress antibody development from months to weeks while improving the specificity and manufacturing properties of new drugs from the start.
The approach enables:
- Precision targeting of disease-causing proteins at vulnerable sites
- Safer therapeutics by avoiding off-target binding that causes side effects
- Rapid pandemic response by quickly generating antibodies against emerging pathogens
- Access to undruggable targets currently inaccessible to existing technologies
Additional information
The lead authors were affiliated with the University of Washington (Nathaniel Bennett, Joseph Watson, Robert Ragotte, Andrew Borst, DéJenaé See, Connor Weidle, and Riti Biswas) and the University of California (Yutong Yu) at the time the research was conducted. Watson, Bennett, and Baker are co-founders of Xaira Therapeutics, which has licensed technology from this study. Study authors Bennett, Watson, Philip Leung, and Buwei Huang are currently employed by Xaira Therapeutics. Study authors Bennett, Watson, Leung, Huang, and current UW Medicine researchers Ragotte, Borst, and Weidle have received payments relating to the licensing of the inventions described in the study to Xaira Therapeutics. All other competing interests are disclosed in the manuscript.
This research was supported by Microsoft, Open Philanthropy, the US National Institutes of Health, and other organizations. All funders are listed in the manuscript.