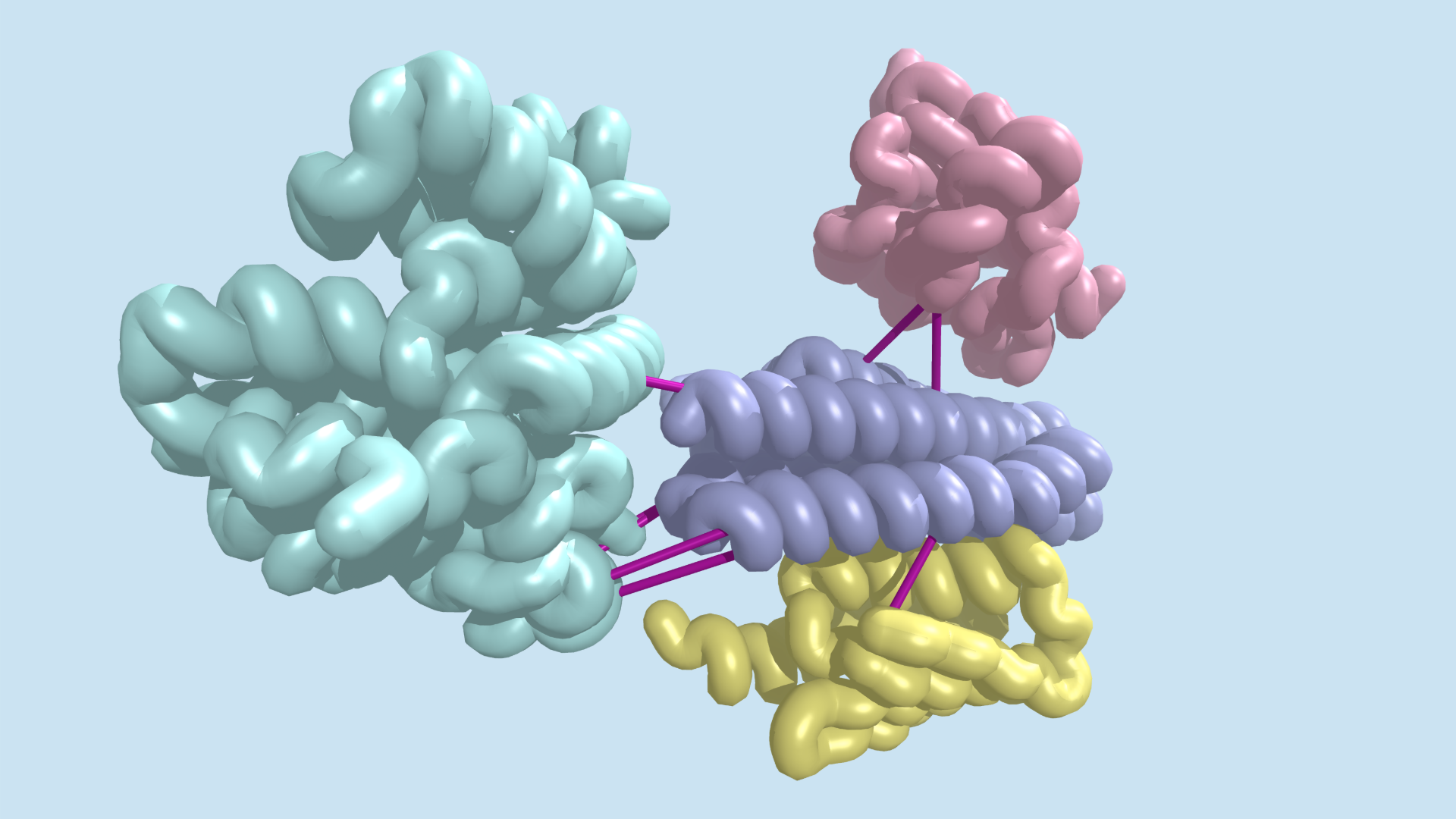
Today we report in Science the identification of hundreds of previously uncharacterized protein–protein interactions in E. coli and the pathogenic bacterium M. tuberculosis. These include both previously unknown protein complexes and previously uncharacterized components of known complexes.
This research was led by postdoctoral fellow Qian Cong and included former Baker lab graduate student Sergey Ovchinnikov, now a John Harvard Distinguished Science Fellow at Harvard.
Augmented by sequences from over 40,000 bacterial genomes, the team assessed coevolution between 5.4 million pairs of E. coliproteins. After finding orthologs and building paired alignments, they used a local statistical model to identify over 21,000 putative protein–protein interactions. Three-dimensional models for proteins in each pair were generated and docked, leading to 804 pairs with the strongest evidence for coevolution.
When compared to predictions inferred from high-throughput experimental screening methods, this new coevolution-based method for identifying protein–protein interactions outperforms in both precision and recall on multiple benchmarks.
814 additional pairs were added to this high-confidence set by incorporating protein pairs reported to interact in experimental studies or on the same operon.
“Coevolution has been useful for understanding how specific proteins interact, but we can now use it as a tool for discovery,” said lead author Qian Cong. “We are going to apply this tool to more pathogens, and the human genome. Our success will depend on how much work other scientists put into annotating which parts of the genome are genes and which parts are something else.”
Read the full report: https://science.sciencemag.org/content/365/6449/185 PDF