We are happy to report that the Rosetta molecular modeling suite was recently used to accurately predict the atomic-scale structure of an important coronavirus protein weeks before it could be measured in the lab. Knowledge gained from studying this viral protein is now being used to guide the design of novel vaccines and antiviral drugs.
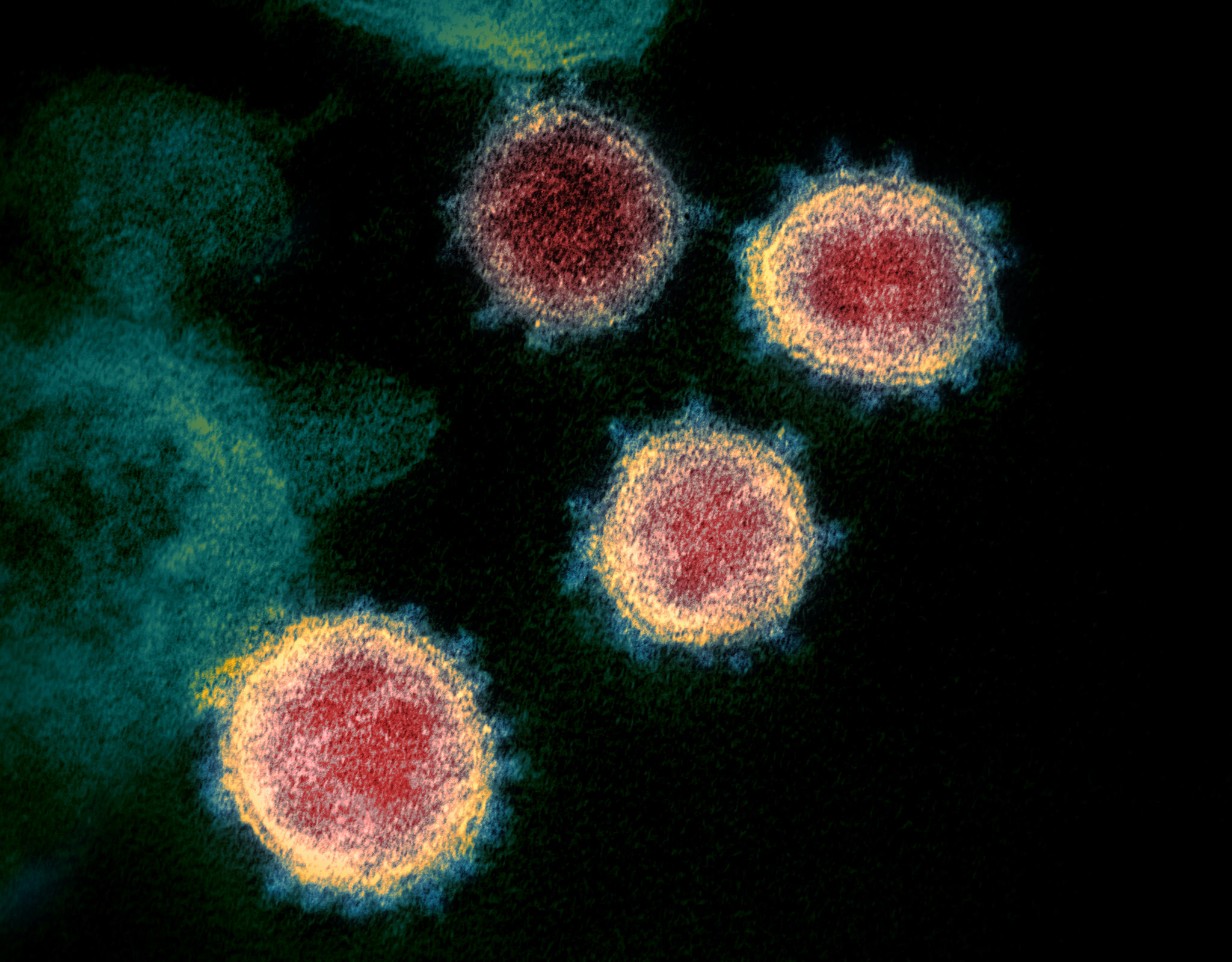
On January 30, the World Health Organization declared the ongoing coronavirus outbreak (COVID-19, caused by the virus SARS-CoV-2) a public health emergency of international concern. Scientists around the world are racing to learn more about this deadly virus which has already spread to more than 30 countries.
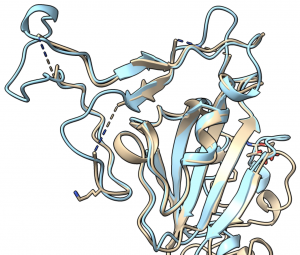
Importantly, structural biologists are quickly gaining insights into what the proteins that make up this virus look like and how they function.
One viral protein in particular — the spike protein — allows SARS-CoV-2 to fuse its membrane with those on human cells, leading to infection. Researchers at UT Austin this week used cryo-electron microscopy to create the first 3D atomic-scale map of the SARS-CoV-2 spike protein in its prefusion state. Like other viral spike proteins, this spear-like molecule is thought to take on two distinct conformations: one before it infects cells, and a different, ‘post-fusion’ state after. Other groups are also applying similar techniques in their laboratories to learn even more about this critically important protein.
Coronavirus spike proteins — like the proteins found in your body — ‘fold up’ in order to function.
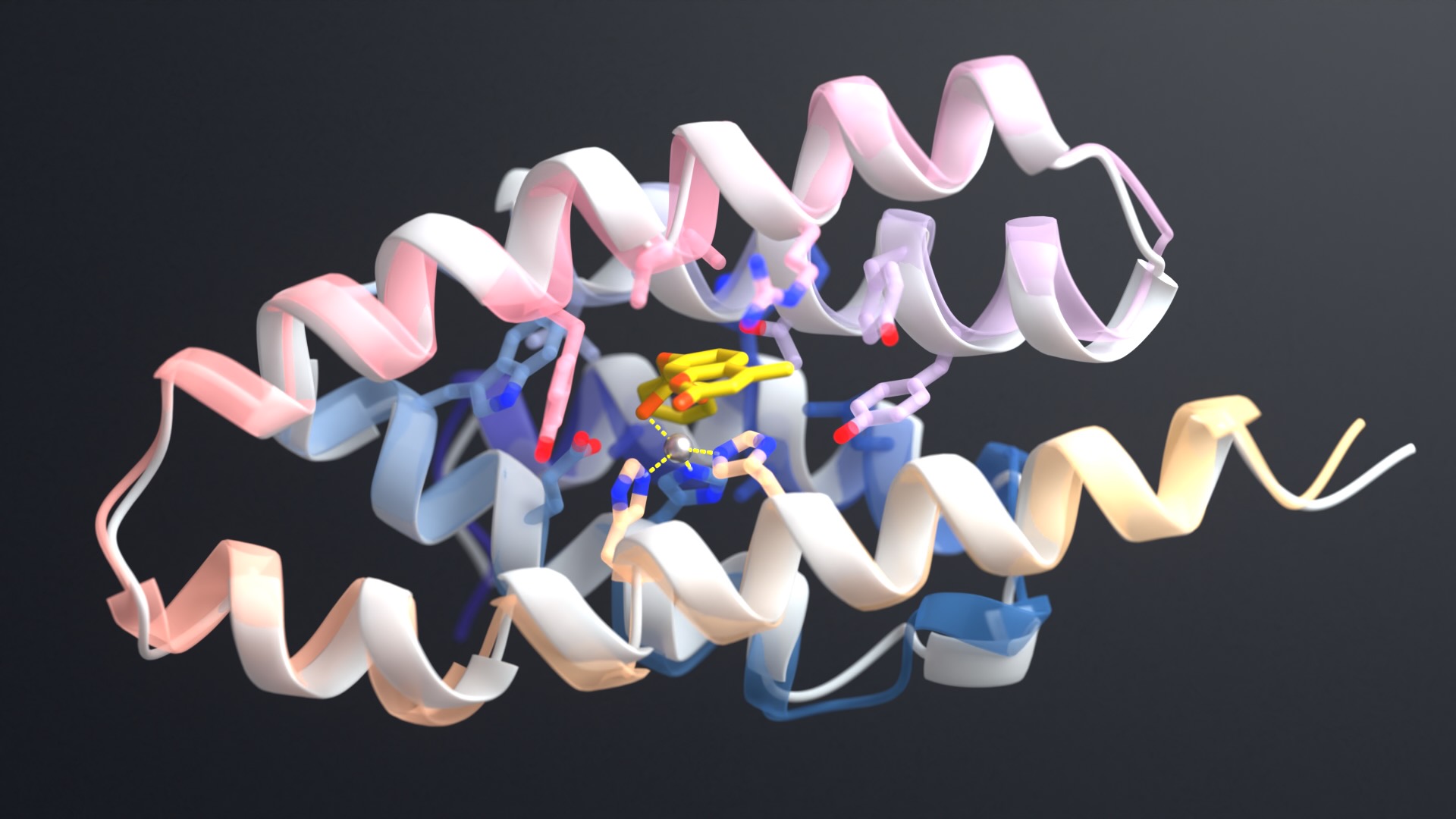
Robetta, our online Rosetta-based protein structure prediction server that is free to use for academics, was able to accurately predict the results of this folding process. In early February, it calculated 3D atomic-scale models of the SARS-CoV-2 spike protein in its prefusion state that closely match those later discovered in the lab.
To download these models, click here.
Designing therapeutics
With this knowledge in hand, researchers at the Institute for Protein Design are now working to create new proteins to neutralize coronavirus. If successful, these antiviral proteins would stick to the SARS-CoV-2 spike protein and thereby prevent viral particles from infecting healthy cells.
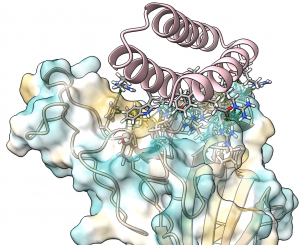
These new drug candidates — a type of molecule we call ‘mini-protein binders’ — seek to combine the specificity of antibodies with the high stability and manufacturability of small molecule drugs. Mini-protein binders are custom-designed on the computer to adhere only to specific targets, such as specific grooves on the SARS-CoV-2 spike protein.
In 2017 we first reported our high-throughput mini-protein binder design strategy. Together with colleagues we designed and tested over 22,000 mini-proteins that target influenza and botulinum neurotoxin B, along with over 6,000 control sequences to probe contributions to folding and binding, and identified 2,618 high-affinity binders.
The de novo designed mini-protein binders produced in that study exhibited much greater stability at elevated temperatures and better neutralization in animal models than comparable antibodies and natural protein derivatives. Probably as a result of their small size and very high stability, they also elicited little immune response. The best of the flu-targeting designs provide prophylactic (before infection) and therapeutic (after infection) protection against influenza infection in mouse models with a potency rivaling or surpassing that of antibodies.
Our researchers are now designing on the computer tens of thousands of anti-coronavirus mini-protein binders. In the coming weeks we hope to produce these mini-proteins in the lab and measure their ability to bind to spike protein. Following this, much more laboratory testing would still be needed to evaluate the safety and efficacy of these experimental coronavirus drugs.
Designing coronavirus vaccines
Technology developed in the King Lab at the Institute for Protein Design is also being applied to try to create an effective vaccine against SARS-CoV-2.
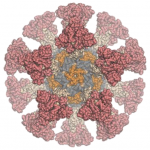
Our colleagues in the Veesler Lab in UW Biochemistry and at the Vaccine Research Center at the National Institutes of Health have fused coronavirus spike proteins to the outside of Rosetta-designed protein nanoparticles to form self-assembling vaccine candidates. Some of these are currently being evaluated in mice. This work builds off our recent efforts to create respiratory vaccines by design.
“We are working with our collaborators at UW, the NIH, and the Bill & Melinda Gates Foundation to help create a safe and effective vaccine for not only SARS-CoV-2 but other coronaviruses as well,” said Neil King, who leads the IPD’s vaccine design efforts.
“This outbreak has illustrated that it’s all hands on deck, and all of us together against the bugs, in the fight against infectious disease. The good news is that the community has developed robust methods for antigen design and display over the last several years that are allowing the rapid generation of vaccine candidates that will likely be highly immunogenic.”