Today we report the development of AI-generated protein catalysts that function efficiently immediately upon synthesis, bypassing the need for lab optimization that has historically bottlenecked enzyme development.
While prior protein design software yielded enzymes with only trace levels of activity, RFdiffusion2 delivers catalytic proficiencies — a rigorous measure of how well an enzyme captures and converts a molecule — comparable to enzymes found in cells.
“Deep learning has evolved from predicting protein structures to creating functional systems. This technology allows us to build enzymes around the specific chemistry we want to see happen, opening the door to advanced medicines and materials that nature has not had time to explore.”
IPD director David Baker, PhD
RFdiffusion2 is freely available on GitHub, allowing all scientists to use and refine this tool.
Publications
Computational design of metallohydrolases
Authors: Donghyo Kim, Seth M. Woodbury, Woody Ahern, Doug Tischer, Alex Kang, Emily Joyce, Asim K. Bera, Nikita Hanikel, Saman Salike, Rohith Krishna, Jason Yim, Samuel J. Pellock, Anna Lauko, Indrek Kalvet, Donald Hilvert, David Baker
Computational design of metallohydrolases
Published in: Nature Methods [PDF]
Authors: Woody Ahern, Jason Yim, Doug Tischer, Saman Salike , Seth M. Woodbury, Donghyo Kim, Indrek Kalvet, Yakov Kipnis , Brian Coventry, Han Raut Altae-Tran, Magnus S. Bauer , Regina Barzilay, Tommi S. Jaakkola, Rohith Krishna, David Baker
Lead Authors




Coverage
Protein design takes big leap forward as model produces enzymes almost as effective as nature’s — Chemistry World
A trio of AI methods tackles enzyme design — C&EN
UW Nobel winner’s lab releases most powerful protein design tool yet — GeekWire
Driving the chemistry of life
Enzymes are the manufacturing machinery of life. Some press atoms together to create larger molecules; others act like nutcrackers, breaking molecules down into smaller components.
For decades, scientists have sought to build custom enzymes for new chemical transformations, such as breaking down plastic pollution. However, efficient catalysts has proven exceptionally difficult to design. Like a nutcracker that must fit a specific shell perfectly to crack it, an enzyme must align with its target molecule with atomic precision. If key features are off by a fraction of a nanometer — the distance a fingernail grows in a single second — the reaction fails.
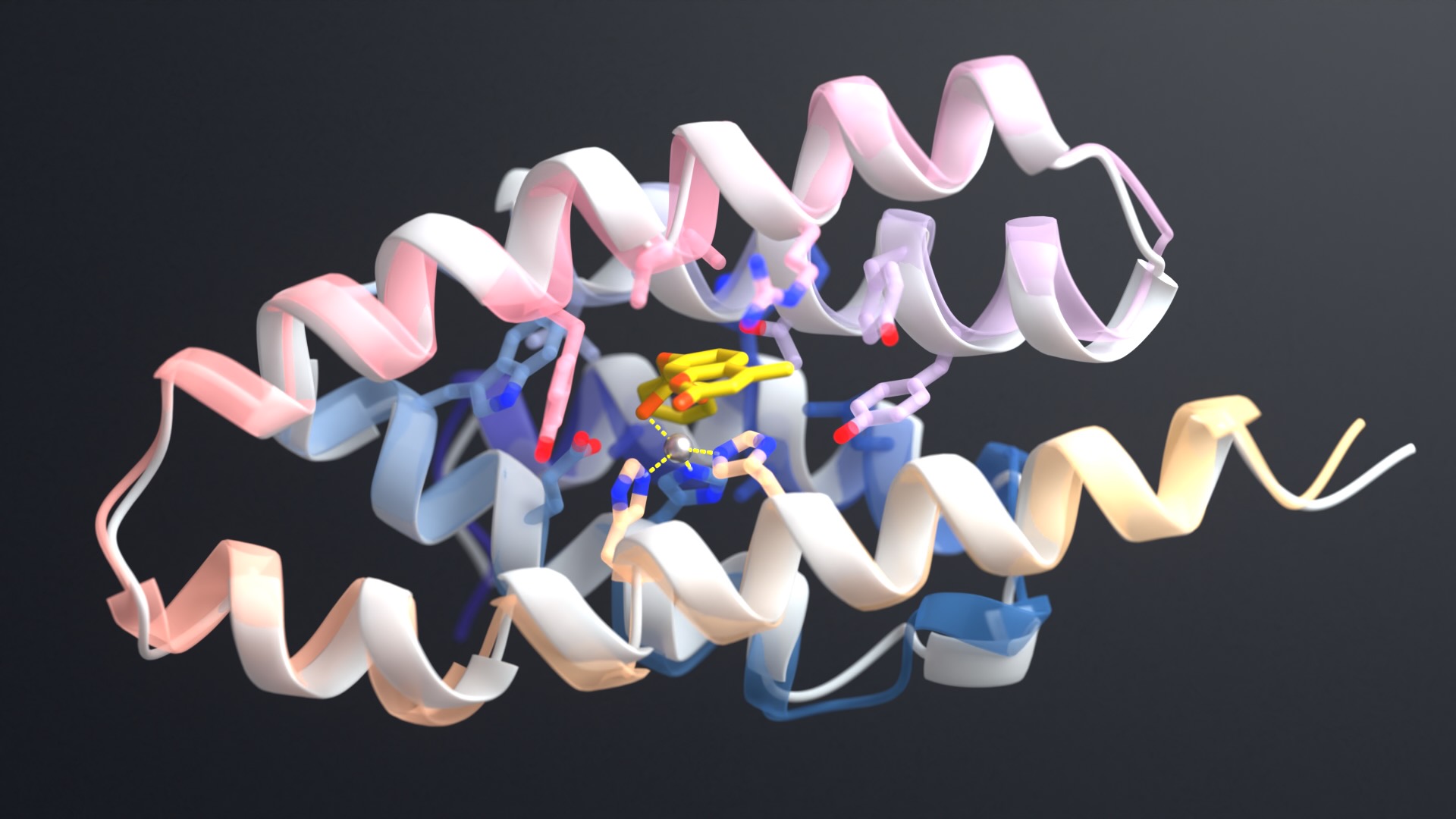
The core innovation of RFdiffusion2 is its ability to treat active sites, or the precise arrangement of atoms within enzymes that drive chemical reactions, as primary input.
Matching natural performance
As reported in Nature, a team led by Baker Lab researchers Donghyo Kim, Seth Woodbury, and Doug Tischer created enzymes unlike any found in nature that cleave chemical bonds. These proteins use a metal ion and activated water molecule to attack target compounds.
Initially, the enzymes generated with RFdiffusion2 showed clear activity, though they operated slower than the most highly evolved natural counterparts. To address this, the team conducted a second round of computational design which yielded much more efficient enzymes, with one achieving levels of catalytic performance common among natural enzymes. X-ray crystallography confirmed that the most active AI-generated enzyme closely matches its computational model, with the protein’s catalytic site precisely positioned as intended — direct evidence of the AI model’s accuracy.
“Historically, computational enzyme design was just the starting point for years of gradual lab optimization. Now, these AI tools give us high-performance catalysts immediately. This shrinks research timelines from months or years into weeks and opens up new possibilities for building catalysts for modern problems, like degrading environmental pollutants.”
Lead author Seth Woodbury
41 out of 41 design challenges solved
In a computational benchmark introduced in the Nature Methods study, the model solved 41 out of 41 difficult enzyme design challenges, compared to just 16 for the previous best protein design tool.
The approach works by providing the AI with only the most important information needed to support a desired chemical reaction, allowing the model to fill in a vast number of important remaining details. Rather than exhaustively searching all possibilities, RFdiffusion2 directly generates valid structures for a given prompt. This, combined with quantum chemistry calculations of reaction mechanisms and deep learning predictions of catalytic features, enables scientists to quickly generate optimal enzymes.
Additional Information
Colleagues across the UW Institute for Protein Design contributed to these projects, along with scientists from the Regina Barzilay Lab and Tommi Jaakkola Lab at MIT and Donald Hilvert Lab at ETH Zurich.
This work was funded by The Audacious Project, Microsoft, Howard Hughes Medical Institute, Open Philanthropy, and the National Institutes of Health, and others. All funders are detailed in the manuscripts.