Latest posts
-
5 questions about designing coronavirus vaccines from our Reddit AMA
Researchers from our vaccine design team recently participated in a Reddit ‘Ask Me Anything’ about our SARS-CoV-2 vaccine research. Reddit users asked over a hundred questions by the time the live event ended — we are sorry we could not address them all. We were lucky to be joined by…
-
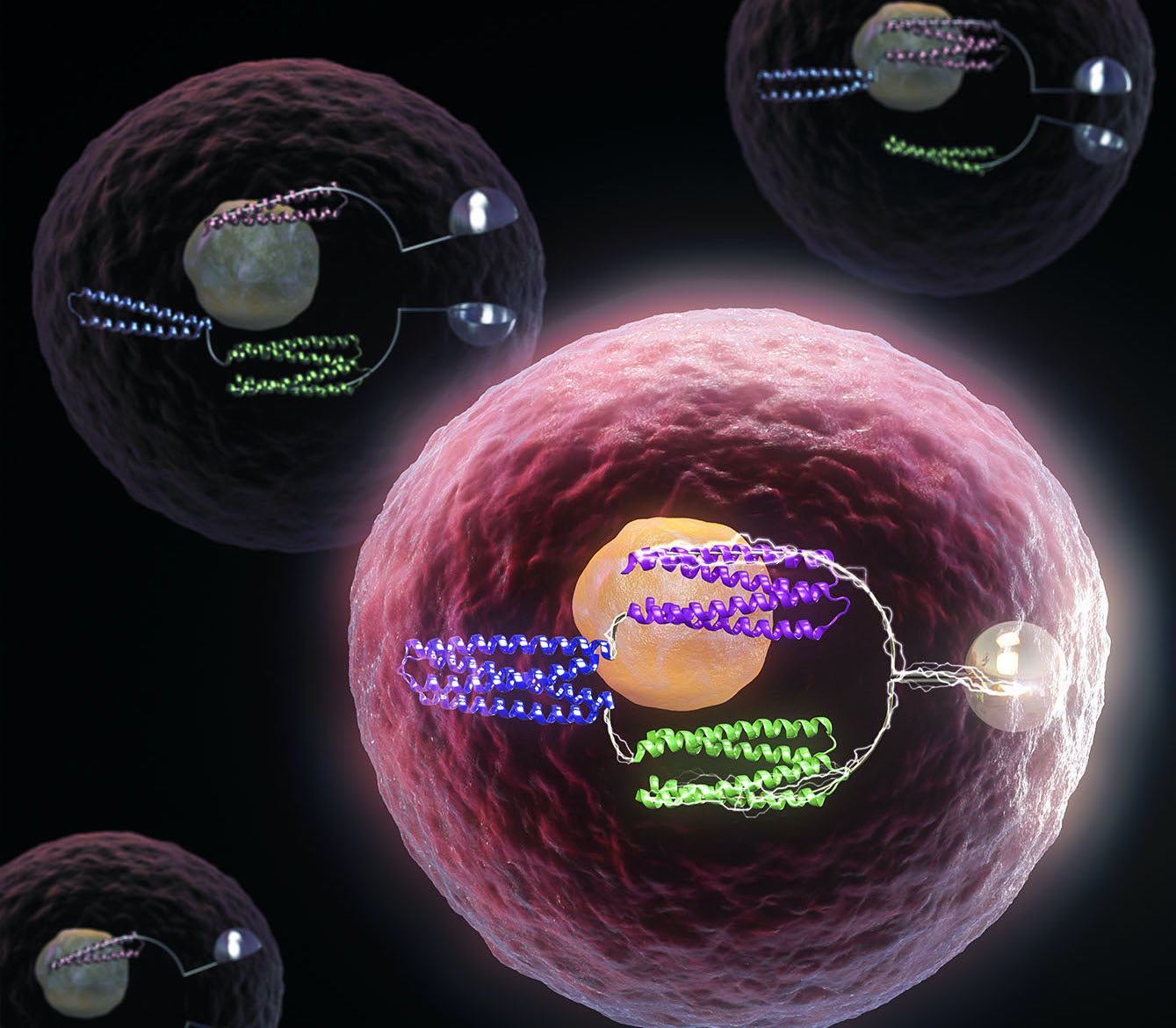
De novo design of protein logic gates
The same basic tools that allow computers to function are now being used to control life at the molecular level, with implications for future medicines and synthetic biology. Together with collaborators, we have created artificial proteins that function as molecular logic gates. These tools, like their electronic counterparts, can be…
-
Volunteers rally to Rosetta@Home to stop COVID-19
As schools, museums, offices and stores shutter to slow the spread of the new coronavirus, millions of people are now finding themselves stuck at home. Fortunately, even in these trying times, there are are small steps that anyone can be take to help combat COVID-19. One option is to donate…
-
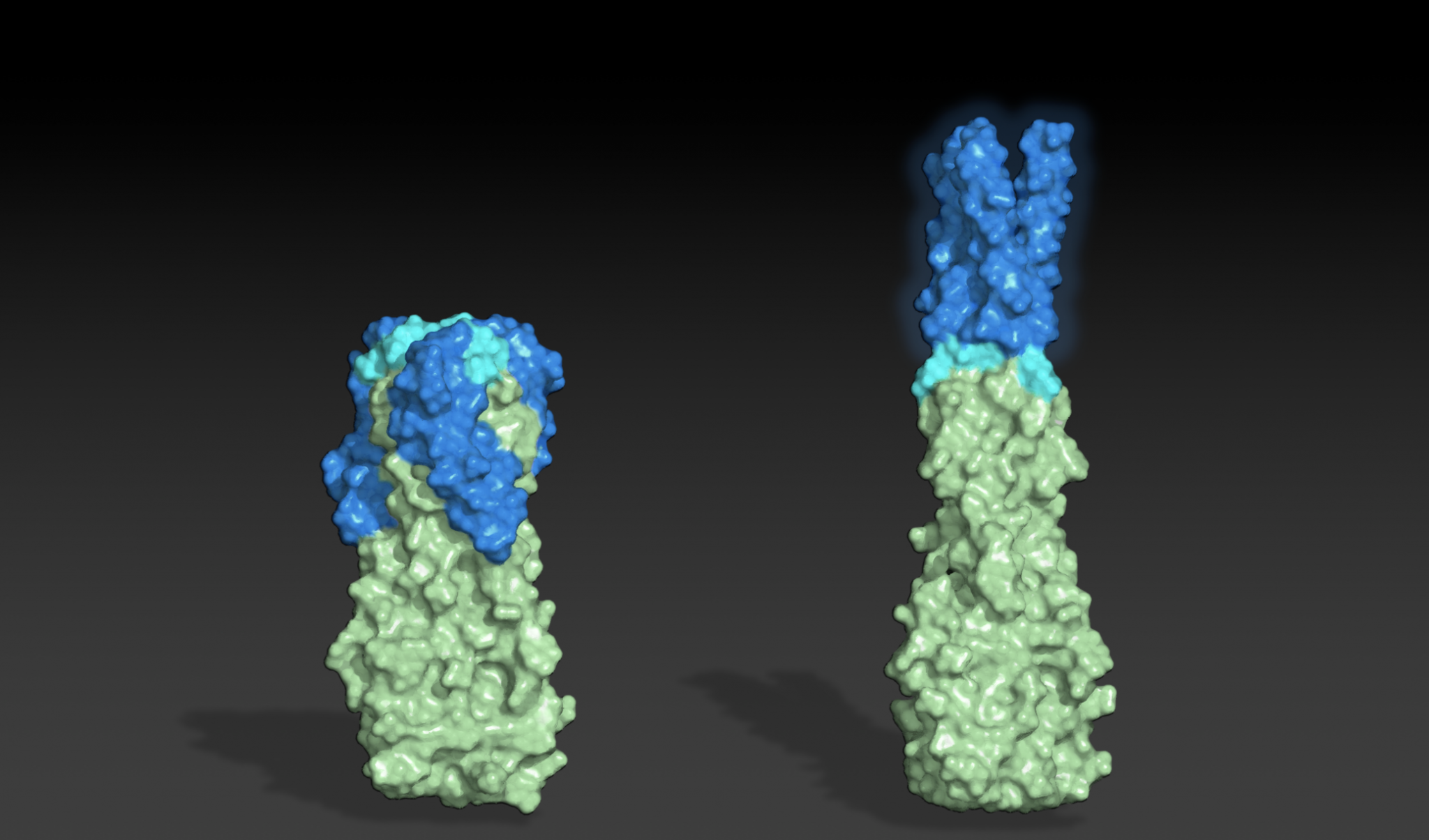
Designing shape-shifting proteins
Today we report the design of protein sequences that adopt more than one well-folded structure, reminiscent of viral fusion proteins. This research moves us closer to creating artificial protein systems with reliable moving parts. In nature, many proteins change shape in response to their environment. This plasticity is often linked…
-
Play Foldit to help stop coronavirus (VIDEO)
You don’t have to be a scientist to do science! By playing the computer game Foldit, you can help discover new antiviral drugs that might stop the novel coronavirus. The most promising solutions will be manufactured and tested at the University of Washington Institute for Protein Design in Seattle. Foldit is run by academic…
-
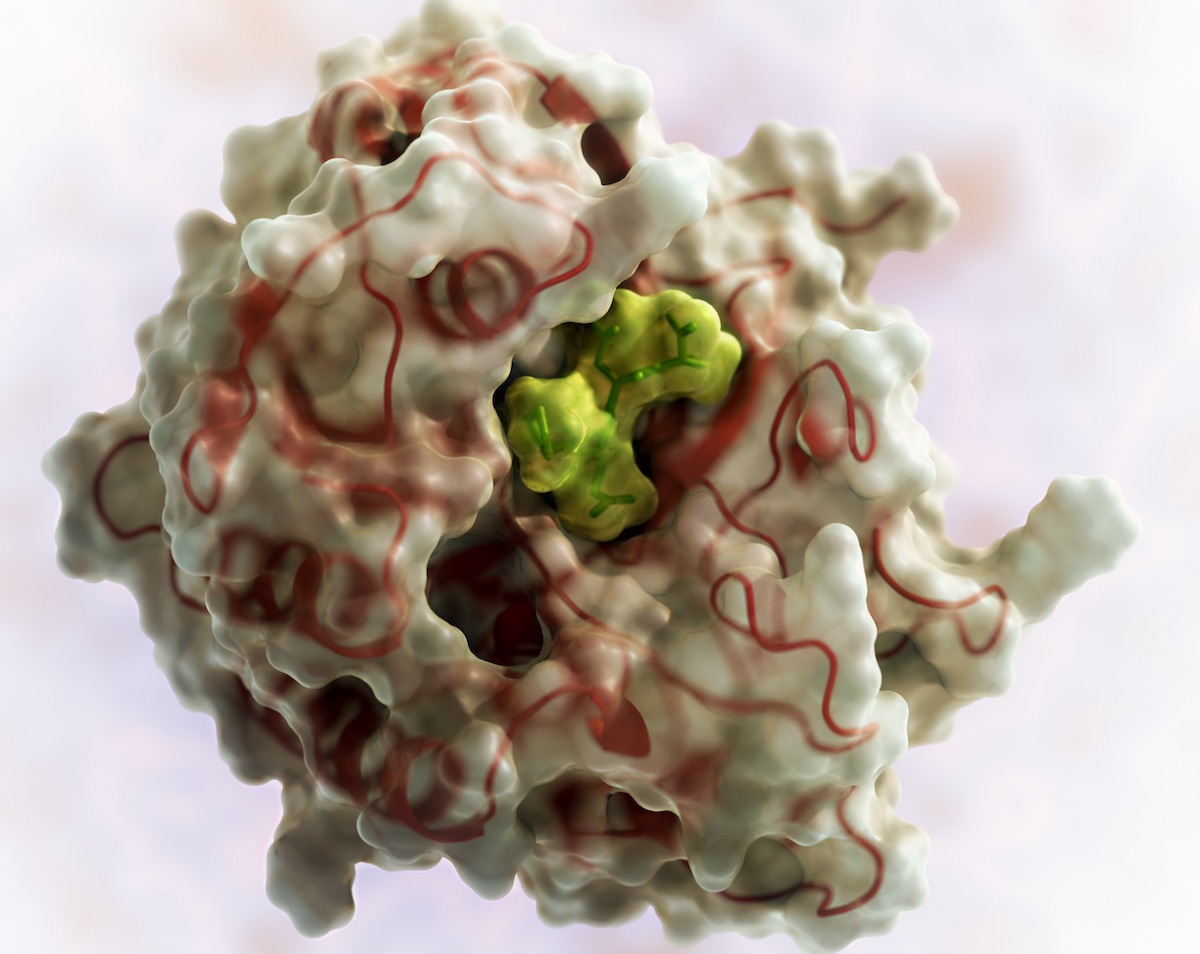
Takeda acquires PvP Biologics
PvP Biologics is on a mission to develop a highly-effective therapeutic to reduce the burden of living with celiac disease. They are advancing an oral enzyme — TAK-062 — designed to break down gluten in the stomach. This exciting research, which began as an iGEM project in 2011, was matured…
-
Rosetta’s role in fighting coronavirus
We are happy to report that the Rosetta molecular modeling suite was recently used to accurately predict the atomic-scale structure of an important coronavirus protein weeks before it could be measured in the lab. Knowledge gained from studying this viral protein is now being used to guide the design of…
-
Introducing WE-REACH, a new biomedical innovation hub
With $4 million in matching funds from the National Institutes of Health, the University of Washington has created a new integrated center to match biomedical discoveries with the resources needed to bring innovative products to the public and improve health. “The University of Washington and regional partner institutions produce some…
-
SCI-STEM Symposium 2020
The Institute for Protein Design at the University of Washington held the second symposium aimed at providing strategies to address diversity challenges in science, technology, engineering, and math (STEM). The Strategies for Cultivating Inclusion in STEM (SCI-STEM) symposium featured leading keynote speakers, panel discussions, and interactive breakout sessions. Members of…
-
Radhika wins poster award at ABRCMS
Undergraduate researcher Radhika Dalal took home a poster award at this year’s Annual Biomedical Research Conference for Minority Students in Anaheim, California. Her mentors include IPD graduate student Una Natterman and Quinton Dowling. Way to go, Radhika! And congratulations to all the award winners: