Latest posts
-
Design of binders for disordered targets
Today we report in Nature the design of proteins that recognize and bind to the so-called “intrinsically disordered regions” of proteins and peptides. The body produces such disordered molecules naturally, but many have been linked to health disorders, including myeloma and other cancers. “Disordered proteins play important roles in biology.…
-
RFdiffusion now free and open source
Today we are making RFdiffusion, our artificial intelligence (AI) program that can generate novel proteins with potential applications in medicine, vaccines, and advanced materials, free for both non-profit and for-profit use under a governed license. The software, which has been tested in our labs, is much faster and more capable…
-
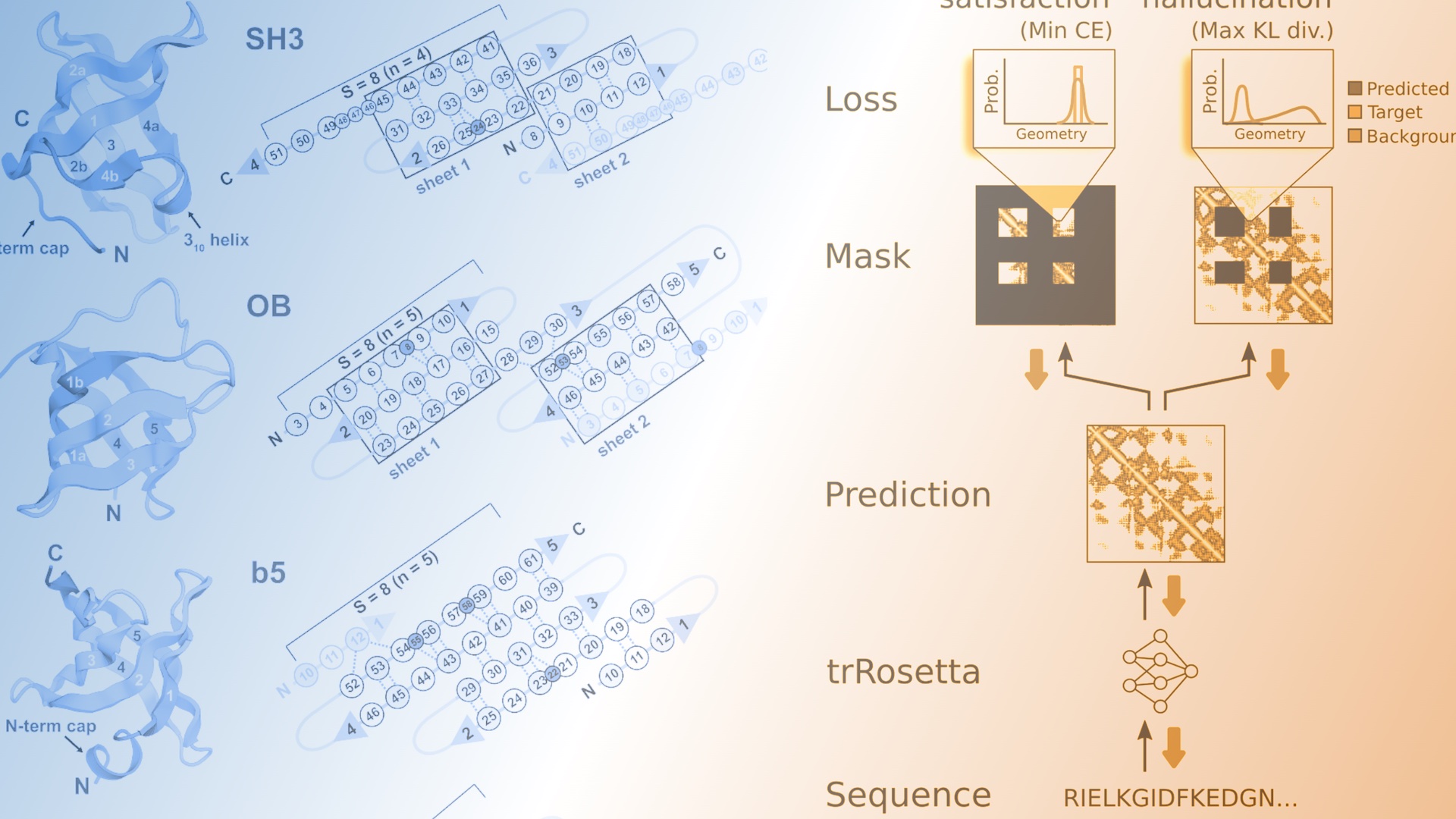
De novo design of small beta barrel proteins
The de novo design of small proteins with beta-barrel topologies has been a challenge for computational design due to the complexity inherent in these folds. In a new study appearing in PNAS, a team led by Baker Lab research scientist David E. Kim describes the successful design and characterization of…
-
Degreaser: Improving protein secretion
King Lab postdoctoral scholars John Wang and Alena Khmelinskaia have developed a new tool for identifying and removing hidden transmembrane sequences that hinder protein secretion.
-
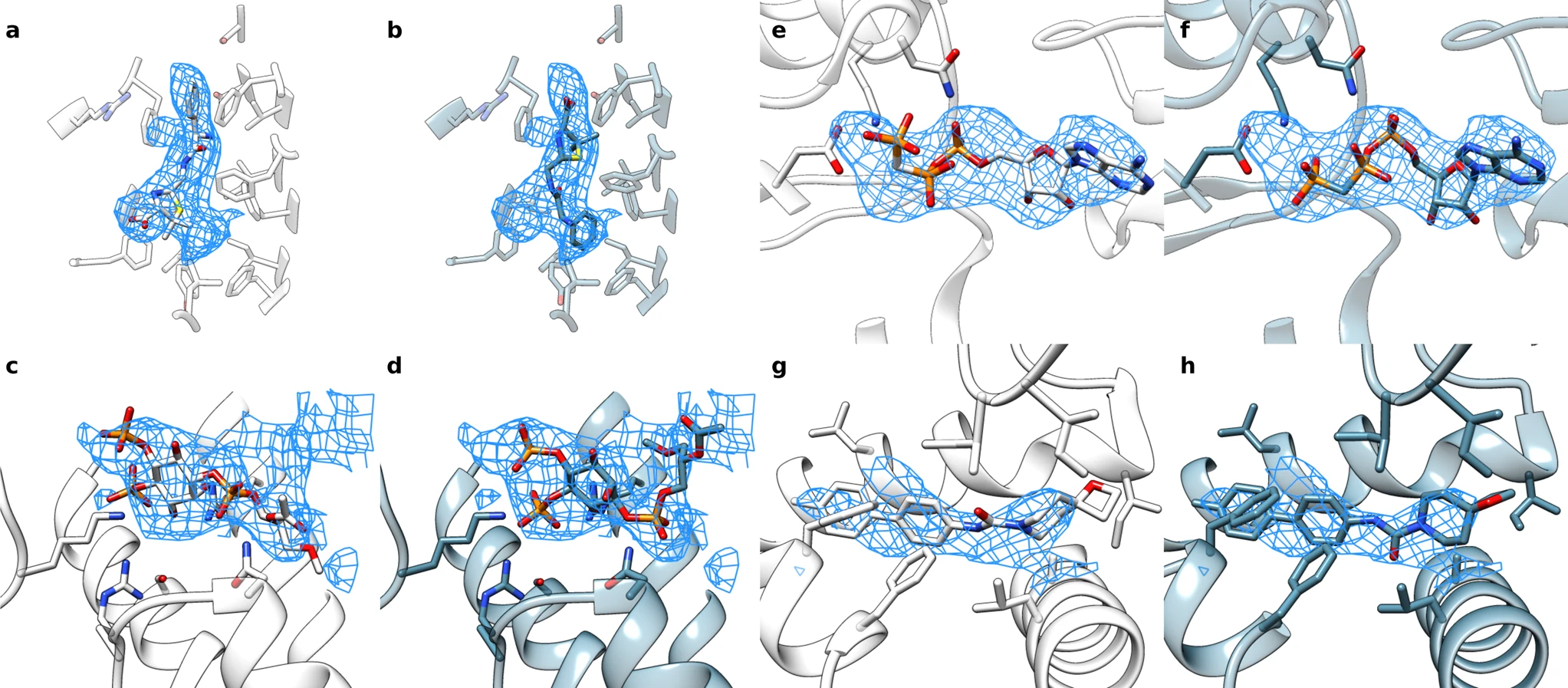
EMERALD: Automatically locate ligands in cyroEM maps
Under ideal conditions, cryo-electron microscopy can be used to determine protein structures at near-atomic resolution. But conditions are not always perfect. To help researchers make use of medium-resolution cryo-electron density maps, scientists in the DiMaio Lab have developed EMERALD, which is a new software tool that can accurately and automatically…
-
Lazarovits and Ueda receive WRF translational funding
Washington Research Foundation (WRF) has awarded a $498,804 technology commercialization grant to Institute for Protein Design researchers Drs. James Lazarovits and George Ueda. The funding will enable Lazarovits and Ueda to further develop their “plug and play” Antibody Cage (AbC) Platform, which converts antibodies into new structures that uniquely bind…
-
Machine learning generates custom enzymes
Today we report in Nature the computational design of highly efficient enzymes unlike any found in nature. Laboratory testing confirms that the new light-emitting enzymes can recognize specific chemical substrates and catalyze the emission of photons very efficiently.
-
NBC: “Scientists use new A.I. tech to fight diseases”
NBC reports on how our scientists are harnessing artificial intelligence to improve how proteins for medicines and vaccines are developed.
-
Frontiers of Knowledge Award goes to David Baker, Demis Hassabis and John Jumper
The 15th BBVA Foundation Frontiers of Knowledge Award in Biology and Biomedicine has gone to David Baker, Demis Hassabis and John Jumper “for their contributions to the use of artificial intelligence for the accurate prediction of the three-dimensional structure of proteins.” From the BBVA Foundation: Baker – a Professor of…
-
NYT: “A.I. Turns Its Artistry to Creating New Human Proteins”
The New York TImes has reported on how, Inspired by digital image generators like DALL-E, our scientists are building artificial intelligences that can fight cancer, flu and COVID-19.